Low affinity glucocorticoid binding site ligands as potential anti-fibrogenics
- PMID: 19432992
- PMCID: PMC2688476
- DOI: 10.1186/1476-5926-8-1
Low affinity glucocorticoid binding site ligands as potential anti-fibrogenics
Abstract
Background: Pregnane X receptor (PXR) agonists inhibit liver fibrosis. However, the rodent PXR activator pregnenolone 16alpha carbonitrile (PCN) blocks, in vitro, hepatic stellate cell-to-myofibroblast trans-differentiation and proliferation in cells from mice with a disrupted PXR gene, suggesting there is an additional anti-fibrogenic drug target for PCN. The role of the low affinity glucocorticoid binding site (LAGS) - which may be identical or associated with the progesterone receptor membrane component 1 (PGRMC1) - in mediating this anti-fibrogenic effect has been examined, since binding of dexamethasone to the LAGS in liver microsomal membranes has previously been shown to be inhibited by PCN.
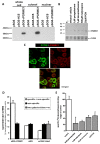
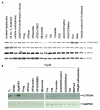
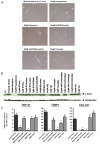
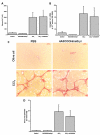
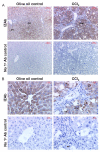
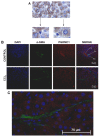
Results: Quiescent rat and human hepatic stellate cells (HSC) were isolated from livers and cultured to generate liver myofibroblasts. HSC and myofibroblasts expressed PGRMC1 as determined by RT-PCR and Western blotting. Quiescent rat HSC also expressed the truncated HC5 variant of rPGRMC1. Rat PGRMC1 was cloned and expression in COS-7 cells gave rise to specific binding of radiolabelled dexamethasone in cell extracts that was inhibited by PCN, suggesting that PGRMC1 may be identical to LAGS or activates LAGS binding activity. Liver microsomes were used to screen a range of structurally related compounds for their ability to inhibit radiolabelled dexamethasone binding to rat LAGS. These compounds were also screened for their ability to activate rat and human PXR and to inhibit rat HSC-to-myofibroblast trans-differentiation/proliferation. A compound (4 androstene-3-one 17beta-carboxylic acid methyl ester) was identified which bound rat LAGS with high affinity and inhibited both rat and human HSC trans-differentiation/proliferation to fibrogenic myofibroblasts without showing evidence of rat or human PXR agonism. However, despite potent anti-fibrogenic effects in vitro, this compound did not modulate liver fibrosis severity in a rat model of liver fibrosis. Immunohistochemical analysis showed that rat liver myofibroblasts in vivo did not express rPGRMC1.
Conclusion: LAGS ligands inhibit HSC trans-differentiation and proliferation in vitro but show little efficacy in inhibiting liver fibrosis, in vivo. The reason(s) for this disparity is/are likely associated with an altered myofibroblast phenotype, in vitro, with expression of rPGMRC1 in vitro but not in vivo. These data emphasize the limitations of in vitro-derived myofibroblasts for predicting their activity in vivo, in studies of fibrogenesis. The data also demonstrate that the anti-fibrogenic effects of PCN in vivo are likely mediated entirely via the PXR.
Figures



Similar articles
-
Pregnenolone-16alpha-carbonitrile inhibits rodent liver fibrogenesis via PXR (pregnane X receptor)-dependent and PXR-independent mechanisms.Biochem J. 2005 May 1;387(Pt 3):601-8. doi: 10.1042/BJ20041598. Biochem J. 2005. PMID: 15595924 Free PMC article.
-
Comparison of the hepatic and thyroid gland effects of sodium phenobarbital in wild type and constitutive androstane receptor (CAR) knockout rats and pregnenolone-16α-carbonitrile in wild type and pregnane X receptor (PXR) knockout rats.Toxicology. 2018 May 1;400-401:20-27. doi: 10.1016/j.tox.2018.03.002. Epub 2018 Mar 13. Toxicology. 2018. PMID: 29548889
-
Disease-induced drug targeting using novel peptide-ligand albumins.J Control Release. 2001 May 14;72(1-3):157-64. doi: 10.1016/s0168-3659(01)00271-1. J Control Release. 2001. PMID: 11389994
-
Comparison of the hepatic and thyroid gland effects of sodium phenobarbital and pregnenolone-16α-carbonitrile in wild-type and constitutive androstane receptor (CAR)/pregnane X receptor (PXR) knockout rats.Xenobiotica. 2019 Feb;49(2):227-238. doi: 10.1080/00498254.2018.1437300. Epub 2018 Mar 26. Xenobiotica. 2019. PMID: 29424600
-
Cooperation of liver cells in health and disease.Adv Anat Embryol Cell Biol. 2001;161:III-XIII, 1-151. doi: 10.1007/978-3-642-56553-3. Adv Anat Embryol Cell Biol. 2001. PMID: 11729749 Review.
Cited by
-
Steroids, Pregnancy and Fetal Development.Front Immunol. 2020 Jan 22;10:3017. doi: 10.3389/fimmu.2019.03017. eCollection 2019. Front Immunol. 2020. PMID: 32038609 Free PMC article. Review.
-
Sustained Isoprostane E2 Elevation, Inflammation and Fibrosis after Acute Ischaemia-Reperfusion Injury Are Reduced by Pregnane X Receptor Activation.PLoS One. 2015 Aug 24;10(8):e0136173. doi: 10.1371/journal.pone.0136173. eCollection 2015. PLoS One. 2015. PMID: 26302150 Free PMC article.
-
Diagnosis and Prediction of CKD Progression by Assessment of Urinary Peptides.J Am Soc Nephrol. 2015 Aug;26(8):1999-2010. doi: 10.1681/ASN.2014050423. Epub 2015 Jan 14. J Am Soc Nephrol. 2015. PMID: 25589610 Free PMC article. Clinical Trial.
-
Utility of B-13 progenitor-derived hepatocytes in hepatotoxicity and genotoxicity studies.Toxicol Sci. 2014 Feb;137(2):350-70. doi: 10.1093/toxsci/kft258. Epub 2013 Nov 14. Toxicol Sci. 2014. PMID: 24235770 Free PMC article.
-
Serine/threonine protein kinase SGK1 in glucocorticoid-dependent transdifferentiation of pancreatic acinar cells to hepatocytes.J Cell Sci. 2011 Feb 1;124(Pt 3):405-13. doi: 10.1242/jcs.077503. Epub 2011 Jan 11. J Cell Sci. 2011. PMID: 21224398 Free PMC article.
References
-
- Fallowfield JA, Mizuno M, Kendall TJ, Constandinou CM, Benyon RC, Duffield JS, Iredale JP. Scar-associated macrophages are a major source of hepatic matrix metalloproteinase-13 and facilitate the resolution of murine hepatic fibrosis. J Immunol. 2007;178:5288–5295. - PubMed
-
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, Perlmann T, Lehmann JM. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/S0092-8674(00)80900-9. - DOI - PubMed
-
- Goodwin B, Gauthier KC, Umetani M, Watson MA, Lochansky MI, Collins L, Leitersdorf E, Mangelsdorf DJ, Kliewer SA, Repa JJ. Identification of bile acid precursors as endogenous ligands for the nuclear xenobiotic pregnane X receptor. Proc Natl Acad Sci USA. 2003;100:223–228. doi: 10.1073/pnas.0237082100. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
