The formation pathway of i-motif tetramers
- PMID: 19433505
- PMCID: PMC2709581
- DOI: 10.1093/nar/gkp340
The formation pathway of i-motif tetramers
Abstract
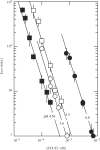
The i-motif is a four-stranded structure formed by two intercalated parallel duplexes containing hemiprotonated C*C(+) pairs. In order to describe the sequence of reactions by which four C-rich strands associate, we measured the formation and dissociation rates of three [TC(n)](4) tetramers (n = 3, 4 and 5), their dissociation constant and the reaction order for tetramer formation by NMR. We find that TC(n) association results in the formation of several tetramers differing by the number of intercalated C*C(+) pairs. The formation rates of the fully and partially intercalated species are comparable but their lifetimes increase strongly with the number of intercalated C*C(+) pairs, and for this reason the single tetramer detected at equilibrium is that with optimal intercalation. The tetramer half formation times vary as the power -2 of the oligonucleotide concentration indicating that the reaction order for i-motif formation is 3. This observation is inconsistent with a model supposing association of two preformed duplex and suggests that quadruplex formation proceeds via sequential strand association into duplex and triplex intermediate species and that triplex formation is rate limiting.
Figures


 and protonated cytidine:
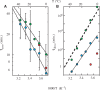
and protonated cytidine:  . (B) Reduced dissociation constant versus pH. The observation that Fi is close 10−4/2 (dashed line) at the temperature for which the association time measured in 10−4 M TC3 solution and the dissociation time are equal shows the consistence of the kinetics and equilibrium measurements.
. (B) Reduced dissociation constant versus pH. The observation that Fi is close 10−4/2 (dashed line) at the temperature for which the association time measured in 10−4 M TC3 solution and the dissociation time are equal shows the consistence of the kinetics and equilibrium measurements.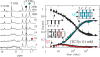
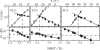

References
-
- Gehring K, Leroy J-L, Guéron M. A tetrameric DNA structure with protonated cytidine-cytidine base pairs. Nature. 1993;363:561–565. - PubMed
-
- Patel D, Bouaziz S, Kettani A, Wang Y. Structure of guanine-rich and cytosine-rich quadruplexes formed in vitro by telomeric, centomeric and triplet repeat disease DNA sequences. In: Neidle S, editor. Oxford Handbook of Nucleic Acid Structure. Oxford University Press; 1999. pp. 389–454.
-
- Leroy JL, Guéron M. Solution structure of the i-motif tetramers of d(TCC), d(5methylCCT) and d(T5methylCC): novel NOE connection between amino protons and sugar protons. Structure. 1995;3:101–120. - PubMed
-
- Nonin S, Phan AT, Leroy J-L. Solution structure and base-pair opening kinetics of the i-motif dimer of d(5mCCTTTACC): a non canonical structure with possible role in chromosome stability. Structure. 1997;5:1231–1246. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

