Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico
- PMID: 19433783
- PMCID: PMC2680428
- DOI: 10.1073/pnas.0903045106
Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico
Abstract
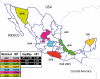
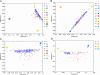
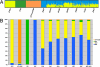
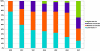
Mexico is developing the basis for genomic medicine to improve healthcare of its population. The extensive study of genetic diversity and linkage disequilibrium structure of different populations has made it possible to develop tagging and imputation strategies to comprehensively analyze common genetic variation in association studies of complex diseases. We assessed the benefit of a Mexican haplotype map to improve identification of genes related to common diseases in the Mexican population. We evaluated genetic diversity, linkage disequilibrium patterns, and extent of haplotype sharing using genomewide data from Mexican Mestizos from regions with different histories of admixture and particular population dynamics. Ancestry was evaluated by including 1 Mexican Amerindian group and data from the HapMap. Our results provide evidence of genetic differences between Mexican subpopulations that should be considered in the design and analysis of association studies of complex diseases. In addition, these results support the notion that a haplotype map of the Mexican Mestizo population can reduce the number of tag SNPs required to characterize common genetic variation in this population. This is one of the first genomewide genotyping efforts of a recently admixed population in Latin America.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Jimenez-Sanchez G. Developing a platform for genomic medicine in Mexico. Science. 2003;300:295–296. - PubMed
-
- Hardy BJ, et al. The next steps for genomic medicine: Challenges and opportunities for the developing world. Nat Rev Genet. 2008;9(Suppl 1):S23–S27. - PubMed
-
- Seguin B, Hardy BJ, Singer PA, Daar AS. Genomics, public health and developing countries: The case of the Mexican National Institute of Genomic Medicine (INMEGEN) Nat Rev Genet. 2008;9(Suppl 1):S5–S9. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

