The adenoviral E1A protein displaces corepressors and relieves gene repression by unliganded thyroid hormone receptors in vivo
- PMID: 19434099
- PMCID: PMC2692372
- DOI: 10.1038/cr.2009.55
The adenoviral E1A protein displaces corepressors and relieves gene repression by unliganded thyroid hormone receptors in vivo
Abstract
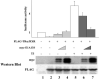
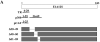
The human adenovirus type 5 early region 1A (E1A) is one of two oncogenes present in the adenovirus genome and functions by interfering with the activities of cellular regulatory proteins. The E1A gene is alternatively spliced to yield five products. Earlier studies have revealed that E1A can regulate the function of thyroid hormone (T3) receptors (TRs). However, analysis in yeast compared with transfection studies in mammalian cell cultures yields surprisingly different effects. Here, we have examined the effect of E1A on TR function by using the frog oocyte in vivo system, where the effects of E1A can be studied in the context of chromatin. We demonstrate that different isoforms of E1A have distinct effects on TR function. The two longest forms inhibit both the repression by unliganded TR and activation by T3-bound TR. We further show that E1A binds to unliganded TR to displace the endogenous corepressor nuclear receptor corepressor, thus relieving the repression by unliganded TR. On the other hand, in the presence of T3, E1A inhibits gene activation by T3-bound TR indirectly, through a mechanism that requires its binding domain for the general coactivator p300. Taken together, our results thus indicate that E1A affects TR function through distinct mechanisms that are dependent upon the presence or absence of T3.
Figures




References
-
- Lazar MA. Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev. 1993;14:184–93. - PubMed
-
- Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Ann Rev Biochem. 1994;63:451–486. - PubMed
-
- Wong J, Shi Y-B. Coordinated regulation of and transcriptional activation by Xenopus thyroid hormone and retinoid X receptors. J Biol Chem. 1995;270:18479–18483. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

