Consistent improvement of cross-docking results using binding site ensembles generated with elastic network normal modes
- PMID: 19434904
- PMCID: PMC2891173
- DOI: 10.1021/ci8003732
Consistent improvement of cross-docking results using binding site ensembles generated with elastic network normal modes
Abstract
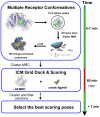
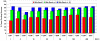
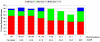
The representation of protein flexibility is still a challenge for the state-of-the-art flexible ligand docking protocols. In this article, we use a large and diverse benchmark to prove that is possible to improve consistently the cross-docking performance against a single receptor conformation, using an equilibrium ensemble of binding site conformers. The benchmark contained 28 proteins, and our method predicted the top-ranked near native ligand poses 20% more efficiently than using a single receptor. The multiple conformations were derived from the collective variable space defined by all heavy-atom elastic network normal modes, including backbone and side chains. We have found that the binding site displacements for best positioning of the ligand seem rather independent from the global collective motions of the protein. We also found that the number of binding site conformations needed to represent nonredundant flexibility was < 100. The ensemble of receptor conformations can be generated at our Web site at http://abagyan.scripps.edu/MRC.
Figures

Similar articles
-
DINC-ensemble: A web server for docking large ligands incrementally to an ensemble of receptor conformations.J Mol Biol. 2025 Aug 1;437(15):169163. doi: 10.1016/j.jmb.2025.169163. Epub 2025 Apr 21. J Mol Biol. 2025. PMID: 40268232
-
Rescoring of docking poses under Occam's Razor: are there simpler solutions?J Comput Aided Mol Des. 2018 Sep;32(9):877-888. doi: 10.1007/s10822-018-0155-5. Epub 2018 Sep 1. J Comput Aided Mol Des. 2018. PMID: 30173397
-
Incorporating backbone flexibility in MedusaDock improves ligand-binding pose prediction in the CSAR2011 docking benchmark.J Chem Inf Model. 2013 Aug 26;53(8):1871-9. doi: 10.1021/ci300478y. Epub 2012 Dec 24. J Chem Inf Model. 2013. PMID: 23237273
-
Pre-existing soft modes of motion uniquely defined by native contact topology facilitate ligand binding to proteins.Protein Sci. 2011 Oct;20(10):1645-58. doi: 10.1002/pro.711. Epub 2011 Sep 9. Protein Sci. 2011. PMID: 21826755 Free PMC article. Review.
-
Principles of flexible protein-protein docking.Proteins. 2008 Nov 1;73(2):271-89. doi: 10.1002/prot.22170. Proteins. 2008. PMID: 18655061 Free PMC article. Review.
Cited by
-
ENCoM server: exploring protein conformational space and the effect of mutations on protein function and stability.Nucleic Acids Res. 2015 Jul 1;43(W1):W395-400. doi: 10.1093/nar/gkv343. Epub 2015 Apr 16. Nucleic Acids Res. 2015. PMID: 25883149 Free PMC article.
-
The anisotropic network model web server at 2015 (ANM 2.0).Bioinformatics. 2015 May 1;31(9):1487-9. doi: 10.1093/bioinformatics/btu847. Epub 2015 Jan 6. Bioinformatics. 2015. PMID: 25568280 Free PMC article.
-
Consensus Induced Fit Docking (cIFD): methodology, validation, and application to the discovery of novel Crm1 inhibitors.J Comput Aided Mol Des. 2012 Nov;26(11):1217-28. doi: 10.1007/s10822-012-9611-9. Epub 2012 Sep 30. J Comput Aided Mol Des. 2012. PMID: 23053738
-
A flexible docking scheme to explore the binding selectivity of PDZ domains.Protein Sci. 2010 May;19(5):914-28. doi: 10.1002/pro.366. Protein Sci. 2010. PMID: 20196074 Free PMC article.
-
Leveraging Fungal and Human Calcineurin-Inhibitor Structures, Biophysical Data, and Dynamics To Design Selective and Nonimmunosuppressive FK506 Analogs.mBio. 2021 Dec 21;12(6):e0300021. doi: 10.1128/mBio.03000-21. Epub 2021 Nov 23. mBio. 2021. PMID: 34809463 Free PMC article.
References
-
- Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. - PubMed
-
- Ishima R, Torchia DA. Protein dynamics from NMR. Nat. Struct. Biol. 2000;7:740–743. - PubMed
-
- Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. - PubMed
-
- Teague SJ. Implications of protein flexibility for drug discovery. Nat. Rev. Drug Dis. 2003;2:527–541. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

