MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents
- PMID: 19435871
- PMCID: PMC4573391
- DOI: 10.1158/1535-7163.MCT-08-1046
MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents
Abstract
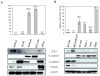
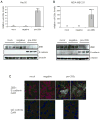
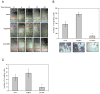
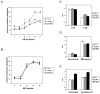
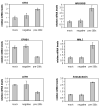
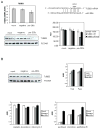
The transcription factor ZEB1 is normally not expressed in epithelial cells. When inappropriately expressed in carcinomas, ZEB1 initiates epithelial to mesenchymal transition due to its ability to repress E-cadherin and other genes involved in polarity. Recently, ZEB1 and ZEB2 have been identified as direct targets of the microRNA-200c family. We find that miR-200c levels are high in well-differentiated endometrial, breast, and ovarian cancer cell lines, but extremely low in poorly differentiated cancer cells. Low or absent miR-200c results in aberrant expression of ZEB1 and consequent repression of E-cadherin. Reinstatement of miR-200c to such cells restores E-cadherin and dramatically reduces migration and invasion. Microarray profiling reveals that in addition to ZEB1 and ZEB2, other mesenchymal genes (such as FN1, NTRK2, and QKI), which are also predicted direct targets of miR-200c, are indeed inhibited by addition of exogenous miR-200c. One such gene, class III β-tubulin (TUBB3), which encodes a tubulin isotype normally found only in neuronal cells, is a direct target of miR-200c. This finding is of particular significance because we show that restoration of miR-200c increases sensitivity to microtubule-targeting agents by 85%. Because expression of TUBB3 is a common mechanism of resistance to microtubule-binding chemotherapeutic agents in many types of solid tumors, the ability of miR-200c to restore chemosensitivity to such agents may be explained by its ability to reduce TUBB3. Because miR-200c is crucial for maintenance of epithelial identity, behavior, and sensitivity to chemotherapy, we propose that it warrants further investigation as a therapeutic strategy for aggressive, drug-resistant cancers.
Figures


References
-
- Singh M, Spoelstra NS, Jean A, et al. ZEB1 expression in type I vs type II endometrial cancers: a marker of aggressive disease. Mod Pathol. 2008;21:912–923. - PubMed
-
- Spoelstra NS, Manning NG, Higashi Y, et al. The transcription factor ZEB1 is aberrantly expressed in aggressive uterine cancers. Cancer Res. 2006;66:3893–3902. - PubMed
-
- Eger A, Aigner K, Sonderegger S, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24:2375–2385. - PubMed
-
- Guaita S, Puig I, Franci C, et al. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem. 2002;277:39209–39216. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

