Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling
- PMID: 19435935
- PMCID: PMC2700536
- DOI: 10.1105/tpc.108.060830
Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling
Abstract
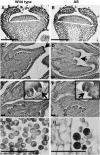
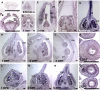
Indole Acetic Acid 9 (IAA9) is a negative auxin response regulator belonging to the Aux/IAA transcription factor gene family whose downregulation triggers fruit set before pollination, thus giving rise to parthenocarpy. In situ hybridization experiments revealed that a tissue-specific gradient of IAA9 expression is established during flower development, the release of which upon pollination triggers the initiation of fruit development. Comparative transcriptome and targeted metabolome analysis uncovered important features of the molecular events underlying pollination-induced and pollination-independent fruit set. Comprehensive transcriptomic profiling identified a high number of genes common to both types of fruit set, among which only a small subset are dependent on IAA9 regulation. The fine-tuning of Aux/IAA and ARF genes and the downregulation of TAG1 and TAGL6 MADS box genes are instrumental in triggering the fruit set program. Auxin and ethylene emerged as the most active signaling hormones involved in the flower-to-fruit transition. However, while these hormones affected only a small number of transcriptional events, dramatic shifts were observed at the metabolic and developmental levels. The activation of photosynthesis and sucrose metabolism-related genes is an integral regulatory component of fruit set process. The combined results allow a far greater comprehension of the regulatory and metabolic events controlling early fruit development both in the presence and absence of pollination/fertilization.
Figures





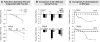


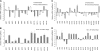



References
-
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251 533–549. - PubMed
-
- Baldet, P., Hernould, M., Laporte, F., Mounet, F., Just, D., Mouras, A., Chevalier, C., and Rothan, C. (2006). The expression of cell proliferation-related genes in early developing flowers is affected by a fruit load reduction in tomato plants. J. Exp. Bot. 57 961–970. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

