Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer
- PMID: 19436038
- PMCID: PMC2684553
- DOI: 10.1093/jnci/djp082
Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer
Abstract
Background: Gene expression profiling of breast cancer has identified two biologically distinct estrogen receptor (ER)-positive subtypes of breast cancer: luminal A and luminal B. Luminal B tumors have higher proliferation and poorer prognosis than luminal A tumors. In this study, we developed a clinically practical immunohistochemistry assay to distinguish luminal B from luminal A tumors and investigated its ability to separate tumors according to breast cancer recurrence-free and disease-specific survival.
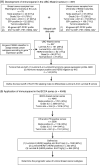
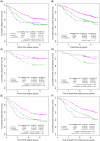
Methods: Tumors from a cohort of 357 patients with invasive breast carcinomas were subtyped by gene expression profile. Hormone receptor status, HER2 status, and the Ki67 index (percentage of Ki67-positive cancer nuclei) were determined immunohistochemically. Receiver operating characteristic curves were used to determine the Ki67 cut point to distinguish luminal B from luminal A tumors. The prognostic value of the immunohistochemical assignment for breast cancer recurrence-free and disease-specific survival was investigated with an independent tissue microarray series of 4046 breast cancers by use of Kaplan-Meier curves and multivariable Cox regression.
Results: Gene expression profiling classified 101 (28%) of the 357 tumors as luminal A and 69 (19%) as luminal B. The best Ki67 index cut point to distinguish luminal B from luminal A tumors was 13.25%. In an independent cohort of 4046 patients with breast cancer, 2847 had hormone receptor-positive tumors. When HER2 immunohistochemistry and the Ki67 index were used to subtype these 2847 tumors, we classified 1530 (59%, 95% confidence interval [CI] = 57% to 61%) as luminal A, 846 (33%, 95% CI = 31% to 34%) as luminal B, and 222 (9%, 95% CI = 7% to 10%) as luminal-HER2 positive. Luminal B and luminal-HER2-positive breast cancers were statistically significantly associated with poor breast cancer recurrence-free and disease-specific survival in all adjuvant systemic treatment categories. Of particular relevance are women who received tamoxifen as their sole adjuvant systemic therapy, among whom the 10-year breast cancer-specific survival was 79% (95% CI = 76% to 83%) for luminal A, 64% (95% CI = 59% to 70%) for luminal B, and 57% (95% CI = 47% to 69%) for luminal-HER2 subtypes.
Conclusion: Expression of ER, progesterone receptor, and HER2 proteins and the Ki67 index appear to distinguish luminal A from luminal B breast cancer subtypes.
Figures



Comment in
-
Re: Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer.J Natl Cancer Inst. 2009 Dec 16;101(24):1730; author reply 1730-1. doi: 10.1093/jnci/djp390. J Natl Cancer Inst. 2009. PMID: 19893007 No abstract available.
References
-
- Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. - PubMed
-
- Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. - PubMed
-
- Tamoxifen for early breast canceran overview of the randomised trials. Early Breast Cancer Trialists’ Collaborative Group. Lancet. 1998;351(9114):1451–1467. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

