Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome
- PMID: 19436716
- PMCID: PMC2675101
- DOI: 10.1371/journal.pgen.1000485
Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome
Abstract
Comparative genomics can be used to infer the history of genomic rearrangements that occurred during the evolution of a species. We used the principle of parsimony, applied to aligned synteny blocks from 11 yeast species, to infer the gene content and gene order that existed in the genome of an extinct ancestral yeast about 100 Mya, immediately before it underwent whole-genome duplication (WGD). The reconstructed ancestral genome contains 4,703 ordered loci on eight chromosomes. The reconstruction is complete except for the subtelomeric regions. We then inferred the series of rearrangement steps that led from this ancestor to the current Saccharomyces cerevisiae genome; relative to the ancestral genome we observe 73 inversions, 66 reciprocal translocations, and five translocations involving telomeres. Some fragile chromosomal sites were reused as evolutionary breakpoints multiple times. We identified 124 genes that have been gained by S. cerevisiae in the time since the WGD, including one that is derived from a hAT family transposon, and 88 ancestral loci at which S. cerevisiae did not retain either of the gene copies that were formed by WGD. Sites of gene gain and evolutionary breakpoints both tend to be associated with tRNA genes and, to a lesser extent, with origins of replication. Many of the gained genes in S. cerevisiae have functions associated with ethanol production, growth in hypoxic environments, or the uptake of alternative nutrient sources.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
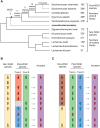


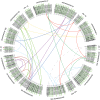


Comment in
-
Reconstructing the history of yeast genomes.PLoS Genet. 2009 May;5(5):e1000483. doi: 10.1371/journal.pgen.1000483. Epub 2009 May 15. PLoS Genet. 2009. PMID: 19436717 Free PMC article. No abstract available.
Similar articles
-
Evolution of gene order in the genomes of two related yeast species.Genome Res. 2001 Dec;11(12):2009-19. doi: 10.1101/gr.212701. Genome Res. 2001. PMID: 11731490
-
The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome.Science. 2004 Apr 9;304(5668):304-7. doi: 10.1126/science.1095781. Epub 2004 Mar 4. Science. 2004. PMID: 15001715
-
Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae.Nature. 2004 Apr 8;428(6983):617-24. doi: 10.1038/nature02424. Epub 2004 Mar 7. Nature. 2004. PMID: 15004568
-
Yeast evolution and comparative genomics.Annu Rev Microbiol. 2005;59:135-53. doi: 10.1146/annurev.micro.59.030804.121400. Annu Rev Microbiol. 2005. PMID: 15877535 Review.
-
Origin, Regulation, and Fitness Effect of Chromosomal Rearrangements in the Yeast Saccharomyces cerevisiae.Int J Mol Sci. 2021 Jan 14;22(2):786. doi: 10.3390/ijms22020786. Int J Mol Sci. 2021. PMID: 33466757 Free PMC article. Review.
Cited by
-
Mitogenomics, phylogeny and morphology reveal two new entomopathogenic species of Ophiocordyceps (Ophiocordycipitaceae, Hypocreales) from south-western China.MycoKeys. 2024 Sep 26;109:49-72. doi: 10.3897/mycokeys.109.124975. eCollection 2024. MycoKeys. 2024. PMID: 39372080 Free PMC article.
-
Comparative gene expression between two yeast species.BMC Genomics. 2013 Jan 16;14:33. doi: 10.1186/1471-2164-14-33. BMC Genomics. 2013. PMID: 23324262 Free PMC article.
-
Genomic instability in an interspecific hybrid of the genus Saccharomyces: a matter of adaptability.Microb Genom. 2020 Oct;6(10):mgen000448. doi: 10.1099/mgen.0.000448. Microb Genom. 2020. PMID: 33021926 Free PMC article.
-
tRNAs as a Driving Force of Genome Evolution in Yeast.Front Microbiol. 2021 Mar 11;12:634004. doi: 10.3389/fmicb.2021.634004. eCollection 2021. Front Microbiol. 2021. PMID: 33776966 Free PMC article. Review.
-
Evolutionary restoration of fertility in an interspecies hybrid yeast, by whole-genome duplication after a failed mating-type switch.PLoS Biol. 2017 May 16;15(5):e2002128. doi: 10.1371/journal.pbio.2002128. eCollection 2017 May. PLoS Biol. 2017. PMID: 28510588 Free PMC article.
References
-
- Keogh RS, Seoighe C, Wolfe KH. Evolution of gene order and chromosome number in Saccharomyces, Kluyveromyces and related fungi. Yeast. 1998;14:443–457. - PubMed
-
- Llorente B, Malpertuy A, Neuveglise C, de Montigny J, Aigle M, et al. Genomic Exploration of the Hemiascomycetous Yeasts: 18. Comparative analysis of chromosome maps and synteny with Saccharomyces cerevisiae. FEBS Lett. 2000;487:101–112. - PubMed
-
- Dujon B. Hemiascomycetous yeasts at the forefront of comparative genomics. Curr Opin Genet Dev. 2005;15:614–620. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

