Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans
- PMID: 19436735
- PMCID: PMC2677459
- DOI: 10.1371/journal.pone.0005517
Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans
Abstract
Background: Mucosa-associated Escherichia coli are frequently found in the colonic mucosa of patients with colorectal adenocarcinoma, but rarely in healthy controls. Chronic mucosal E. coli infection has therefore been linked to colonic tumourigenesis. E. coli strains carrying eae (encoding the bacterial adhesion protein intimin) attach intimately to the intestinal mucosa and are classed as attaching and effacing E. coli (AEEC). Enteropathogenic Escherichia coli (EPEC) are the most common form of AEEC identified in man. EPEC utilise a type III secretion system to translocate effector proteins into host cells and infection induces wide-ranging effects on the host cell proteome. We hypothesised that EPEC infection could influence molecular pathways involved in colorectal tumourigenesis.
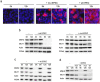
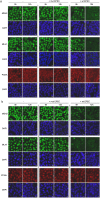
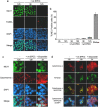
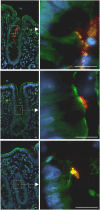
Methodology/principal findings: When co-cultured with human colorectal cell lines, EPEC dramatically downregulated the expression of key DNA mismatch repair proteins MSH2 and MLH1 in an attachment specific manner. Cytochrome c staining and TUNEL analysis confirmed that this effect was not a consequence of apoptosis/necrosis. Ex vivo human colonic mucosa was co-cultured with EPEC and probed by immunofluorescence to locate adherent bacteria. EPEC entered 10% of colonic crypts and adhered to crypt epithelial cells, often in the proliferative compartment. Adenocarcinoma and normal colonic mucosa from colorectal cancer patients (n = 20) was probed by immunofluorescence and PCR for AEEC. Mucosa-associated E. coli were found on 10/20 (50%) adenocarcinomas and 3/20 (15%) normal mucosa samples (P<0.05). AEEC were detected on 5/20 (25%) adenocarcinomas, but not normal mucosa samples (P<0.05).
Significance/conclusions: The ability of EPEC to downregulate DNA mismatch repair proteins represents a novel gene-environment interaction that could increase the susceptibility of colonic epithelial cells to mutations and therefore promote colonic tumourigenesis. The potential role of AEEC in colorectal tumourigenesis warrants further investigation.
Conflict of interest statement
Figures


References
-
- Stewart BW, Kleihues P, editors. World Cancer Report. Lyon: World health Organisation, International Agency for Research on Cancer, IARC Press; 2003. Colorectal Cancer. p. 198.
-
- de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769–780. - PubMed
-
- Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127:80–93. - PubMed
-
- Swidsinski A, Khilkin M, Kerjaschki D, Schreiber S, Ortner M, et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115:281–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

