Developing bifunctional beta-lactamase molecules with built-in target-recognizing module for prodrug therapy: identification of Enterobacter Cloacae P99 cephalosporinase loops suitable for randomization and phage-display selection
- PMID: 19437416
- PMCID: PMC2824592
- DOI: 10.1002/jmr.957
Developing bifunctional beta-lactamase molecules with built-in target-recognizing module for prodrug therapy: identification of Enterobacter Cloacae P99 cephalosporinase loops suitable for randomization and phage-display selection
Abstract
This study was focused on developing catalytically active beta-lactamase enzyme molecules that have target-recognizing sites built within their scaffold. Using phage-display approach, nine libraries were constructed by inserting the randomized linear or cysteine-constrained heptapeptides in the five different loops on the outer surface of P99 beta-lactamase molecule. The pIII signal peptide of Sec-pathway was employed for a periplasmic translocation of the beta-lactamase fusion protein, which we found more efficient than the DsbA signal peptide of SRP-pathway. The randomized heptapeptide loops replaced native amino acids between positions (34)Y-(37)K, (238)M-(246)A, (275)N-(280)A, (305)A-(311)S, or (329)I-(334)I of the P99 beta-lactamase molecules for generating the loop-1 to -5 libraries, respectively. The diversity of each loop library was judged by counting the primary and beta-lactamase-active clones. The linear peptide inserts in the loop-2 library showed the maximum number of the beta-lactamase-active clones, followed by the loop-5, loop-3, and loop-4. The insertion of the cysteine-constrained loops exhibited a dramatic loss of the enzyme-active beta-lactamase clones. The complexity of the loop-2 linear library, as determined by the frequency and diversity of amino acid distributions in the randomized region, appears consistent with the standards of other types of phage display library systems. The selection of the loop-2 linear library on streptavidin protein as a test target identified several beta-lactamase clones that specifically bound to streptavidin. In conclusion, this study identified the suitability of the loop-2 of P99 beta-lactamase for constructing a phage-display library of the beta-lactamase enzyme-active molecules that can be selected against a target. This is an enabling step in our long-term goal of developing bifunctional beta-lactamase molecules against cancer-specific targets for enzyme prodrug therapy of cancer.
Figures

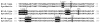


References
-
- Bagshawe KD. Antibody-directed enzyme prodrug therapy (ADEPT) for cancer. Expert Rev Anticancer Ther. 2006;6:1421–1431. - PubMed
-
- Bhatia J, Sharma SK, Chester KA, Pedley RB, Boden RW, Read DA, Boxer GM, Michael NP, Begent RH. Catalytic activity of an in vivo tumor targeted anti-CEA scFv::carboxypeptidase G2 fusion protein. Int J Cancer. 2000;85:571–577. - PubMed
-
- Cantu C, 3rd, Huang W, Palzkill T. Cephalosporin substrate specificity determinants of TEM-1 beta-lactamase. J Biol Chem. 1997;272:29144–29150. - PubMed
-
- Cortez-Retamozo V, Backmann N, Senter PD, Wernery U, De Baetselier P, Muyldermans S, Revets H. Efficient cancer therapy with a nanobody-based conjugate. Cancer Res. 2004;64:2853–2857. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

