Features distinguishing Epstein-Barr virus infections of epithelial cells and B cells: viral genome expression, genome maintenance, and genome amplification
- PMID: 19439479
- PMCID: PMC2708605
- DOI: 10.1128/JVI.00108-09
Features distinguishing Epstein-Barr virus infections of epithelial cells and B cells: viral genome expression, genome maintenance, and genome amplification
Abstract
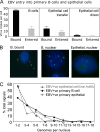
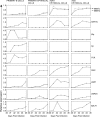
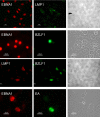
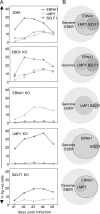
Epstein-Barr virus (EBV) is associated with malignant diseases of lymphoid and epithelial cell origin. The tropism of EBV is due to B-cell-restricted expression of CD21, the major receptor molecule for the virus. However, efficient infection of CD21- epithelial cells can be achieved via transfer from EBV-coated B cells. We compare and contrast here the early events following in vitro infection of primary B cells and epithelial cells. Using sensitive, quantitative reverse transcription-PCR assays for several latent and lytic transcripts and two-color immunofluorescence staining to analyze expression at the single cell level, we confirmed and extended previous reports indicating that the two cell types support different patterns of transcription. Furthermore, whereas infection of B cells with one or two copies of EBV resulted in rapid amplification of the viral genome to >20 copies per cell, such amplification was not normally observed after infection of primary epithelial cells or undifferentiated epithelial lines. In epithelial cells, EBNA1 expression was detected in only ca. 40% of EBER+ cells, and the EBV genome was subsequently lost during prolonged culture. One exception was that infection of AGS, a gastric carcinoma line, resulted in maintenance of EBNA1 expression and amplification of the EBV episome. In contrast to B cells, where amplification of the EBV episome occurred even with a replication-defective BZLF1-knockout virus, amplification in AGS cells was dependent upon early lytic cycle gene expression. These data highlight the influence of the host cell on the outcome of EBV infection with regard to genome expression, amplification, and maintenance.
Figures


References
-
- Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9395-404. - PubMed
-
- Becker, J., U. Leser, M. Marschall, A. Langford, W. Jilg, H. Gelderblom, P. Reichart, and H. Wolf. 1991. Expression of proteins encoded by Epstein-Barr virus transactivator genes depends on the differentiation of epithelial cells in oral hairy leukoplakia. Proc. Natl. Acad. Sci. USA 888332-8336. - PMC - PubMed
-
- Bell, A. I., K. Groves, G. L. Kelly, D. Croom-Carter, E. Hui, A. T. Chan, and A. B. Rickinson. 2006. Analysis of Epstein-Barr virus latent gene expression in endemic Burkitt's lymphoma and nasopharyngeal carcinoma tumour cells by using quantitative real-time PCR assays. J. Gen. Virol. 872885-2890. - PubMed
-
- Busson, P., Q. Zhang, J. M. Guillon, C. D. Gregory, L. S. Young, B. Clausse, M. Lipinski, A. B. Rickinson, and T. Tursz. 1992. Elevated expression of ICAM1 (CD54) and minimal expression of LFA3 (CD58) in Epstein-Barr-virus-positive nasopharyngeal carcinoma cells. Int. J. Cancer 50863-867. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

