MicroRNA-127 modulates fetal lung development
- PMID: 19439715
- PMCID: PMC2685501
- DOI: 10.1152/physiolgenomics.90268.2008
MicroRNA-127 modulates fetal lung development
Abstract
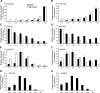
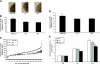
MicroRNAs (miRNAs) are small endogenous RNAs and are widely regarded as one of the most important regulators of gene expression in both plants and animals. To define the roles of miRNAs in fetal lung development, we profiled the miRNA expression pattern during lung development with a miRNA microarray. We identified 21 miRNAs that showed significant changes in expression during lung development. These miRNAs were grouped into four distinct clusters based on their expression pattern. Cluster 1 contained miRNAs whose expression increased as development progressed, while clusters 2 and 3 showed the opposite trend of expression. miRNAs in cluster 4 including miRNA-127 (miR-127) had the highest expression at the late stage of fetal lung development. Quantitative real-time PCR validated the microarray results of six selected miRNAs. In situ hybridization demonstrated that miR-127 expression gradually shifted from mesenchymal cells to epithelial cells as development progressed. Overexpression of miR-127 in fetal lung organ culture significantly decreased the terminal bud count, increased terminal and internal bud sizes, and caused unevenness in bud sizes, indicating improper development. These findings suggest that miR-127 may have an important role in fetal lung development.
Figures

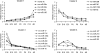


References
-
- Bartel DP MicroRNAs: genomics, biogenesis, mechanism, function. Cell 116: 281–297, 2004. - PubMed
-
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36, 2003. - PubMed
-
- Burri PH Fetal and postnatal development of the lung. Annu Rev Physiol 46: 617–628, 1984. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

