Identification of new SLE-associated genes with a two-step Bayesian study design
- PMID: 19440200
- PMCID: PMC3434884
- DOI: 10.1038/gene.2009.38
Identification of new SLE-associated genes with a two-step Bayesian study design
Abstract
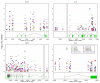
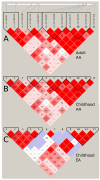
In our earlier study, we utilized a Bayesian design to probe the association of approximately 1000 genes (approximately 10,000 single-nucleotide polymorphisms (SNPs)) with systemic lupus erythematosus (SLE) on a moderate number of trios of parents and children with SLE. Two genes associated with SLE, with a multitest-corrected false discovery rate (FDR) of <0.05, were identified, and a number of noteworthy genes with FDR of <0.8 were also found, pointing out a future direction for the study. In this report, using a large population of controls and adult- or childhood-onset SLE cases, we have extended the earlier investigation to explore the SLE association of 10 of these noteworthy genes (109 SNPs). We have found that seven of these genes exhibit a significant (FDR<0.05) association with SLE, both confirming some genes that have earlier been found to be associated with SLE (PTPN22 and IRF5) and presenting novel findings of genes (KLRG1, interleukin-16, protein tyrosine phosphatase receptor type T, toll-like receptor (TLR)8 and CASP10), which have not been reported earlier. The results signify that the two-step candidate pathway design is an efficient way to study the genetic foundations of complex diseases. Furthermore, the novel genes identified in this study point to new directions in both the diagnosis and the eventual treatment of this debilitating disease.
Figures

References
-
- Collins FS, Guyer MS, Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. - PubMed
-
- Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–1344. - PubMed
-
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. - PubMed
Publication types
MeSH terms
Grants and funding
- AR049084/AR/NIAMS NIH HHS/United States
- P60 AR048095/AR/NIAMS NIH HHS/United States
- P20 RR020143/RR/NCRR NIH HHS/United States
- AR42460/AR/NIAMS NIH HHS/United States
- P30 AR053483/AR/NIAMS NIH HHS/United States
- AR053483/AR/NIAMS NIH HHS/United States
- R01 AI024717/AI/NIAID NIH HHS/United States
- R01 AR045650/AR/NIAMS NIH HHS/United States
- AR048940/AR/NIAMS NIH HHS/United States
- AR043274/AR/NIAMS NIH HHS/United States
- R01 AR043274/AR/NIAMS NIH HHS/United States
- R01AR445650/AR/NIAMS NIH HHS/United States
- RR020143/RR/NCRR NIH HHS/United States
- AI24717/AI/NIAID NIH HHS/United States
- R37 AI024717/AI/NIAID NIH HHS/United States
- R01 AR042460/AR/NIAMS NIH HHS/United States
- R01 AR057172/AR/NIAMS NIH HHS/United States
- AI063622/AI/NIAID NIH HHS/United States
- P01-AR49084/AR/NIAMS NIH HHS/United States
- P60-AR48095/AR/NIAMS NIH HHS/United States
- AR445650/AR/NIAMS NIH HHS/United States
- P50 AR048940/AR/NIAMS NIH HHS/United States
- P01 AR049084/AR/NIAMS NIH HHS/United States
- R01 AI063622/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

