Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli
- PMID: 19442264
- PMCID: PMC2689190
- DOI: 10.1186/1475-2859-8-26
Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli
Abstract
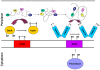
Bacteria are simple and cost effective hosts for producing recombinant proteins. However, their physiological features may limit their use for obtaining in native form proteins of some specific structural classes, such as for instance polypeptides that undergo extensive post-translational modifications. To some extent, also the production of proteins that depending on disulfide bridges for their stability has been considered difficult in E. coli.Both eukaryotic and prokaryotic organisms keep their cytoplasm reduced and, consequently, disulfide bond formation is impaired in this subcellular compartment. Disulfide bridges can stabilize protein structure and are often present in high abundance in secreted proteins. In eukaryotic cells such bonds are formed in the oxidizing environment of endoplasmic reticulum during the export process. Bacteria do not possess a similar specialized subcellular compartment, but they have both export systems and enzymatic activities aimed at the formation and at the quality control of disulfide bonds in the oxidizing periplasm.This article reviews the available strategies for exploiting the physiological mechanisms of bactera to produce properly folded disulfide-bonded proteins.
Figures

References
-
- Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–135. - PubMed
-
- Messens J, Collet J-F. Pathways of disulfide bond formation in Escherichia coli. Int J Biochem Cell Biol. 2006;38:1050–1062. - PubMed
-
- Berndt C, Lillig CH, Holmgren A. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim Biophys Acta. 2008;1783:641–650. - PubMed
-
- Mavrangelos C, Thiel M, Adamson PJ, Millard DJ, Nobbs S, Zola H, Nicholson IC. Increased yield and activity of soluble single-chain antibody fragments by combining high-level expression and the Skp peripasmic chaperonin. Protein Expr Purif. 2001;23:289–295. - PubMed
-
- Simmons LC, Yansura DG. Translational level is a critical factor for the secretion of the heterologous proteins in Escherichia coli. Nat Biotechnol. 14:629–634. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

