Structural insights into RNA splicing
- PMID: 19443210
- PMCID: PMC2756803
- DOI: 10.1016/j.sbi.2009.04.002
Structural insights into RNA splicing
Abstract
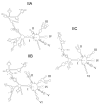
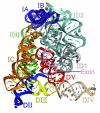
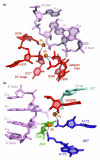
Intron splicing is a fundamental biological process whereby noncoding sequences are removed from precursor RNAs. Recent work has provided new insights into the structural features and reaction mechanisms of two introns that catalyze their own splicing from precursor RNA: the group I and II introns. In addition, there is an increasing amount of structural information on the spliceosome, which is a ribonucleoprotein machine that catalyzes nuclear pre-mRNA splicing in eukaryotes. Here, we compare structures and catalytic mechanisms of self-splicing RNAs and we discuss the possible implications for spliceosomal reaction mechanisms.
Figures
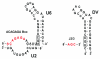
References
-
- Vicens Q, Cech T. Atomic level architecture of group I introns revealed. Trends Biochem Sci. 2006;31:41–51. - PubMed
-
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. - PubMed
-
- Peebles C, Perlman P, Mecklenburg K, Petrillo M, Tabor J, Jarrell K, Cheng H. A self-splicing RNA excises an intron lariat. Cell. 1986;44:213–223. - PubMed
-
- Pyle A, Fedorova O, Waldsich C. Folding of group II introns: a model system for large, multidomain RNAs? Trends Biochem Sci. 2007;32:138–145. - PubMed
-
- Michel F, Ferat J. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

