MM-align: a quick algorithm for aligning multiple-chain protein complex structures using iterative dynamic programming
- PMID: 19443443
- PMCID: PMC2699532
- DOI: 10.1093/nar/gkp318
MM-align: a quick algorithm for aligning multiple-chain protein complex structures using iterative dynamic programming
Abstract
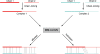
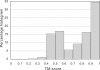
Structural comparison of multiple-chain protein complexes is essential in many studies of protein-protein interactions. We develop a new algorithm, MM-align, for sequence-independent alignment of protein complex structures. The algorithm is built on a heuristic iteration of a modified Needleman-Wunsch dynamic programming (DP) algorithm, with the alignment score specified by the inter-complex residue distances. The multiple chains in each complex are first joined, in every possible order, and then simultaneously aligned with cross-chain alignments prevented. The alignments of interface residues are enhanced by an interface-specific weighting factor. MM-align is tested on a large-scale benchmark set of 205 x 3897 non-homologous multiple-chain complex pairs. Compared with a naïve extension of the monomer alignment program of TM-align, the alignment accuracy of MM-align is significantly higher as judged by the average TM-score of the physically-aligned residues. MM-align is about two times faster than TM-align because of omitting the cross-alignment zone of the DP matrix. It also shows that the enhanced alignment of the interfaces helps in identifying biologically relevant protein complex pairs.
Figures


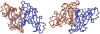
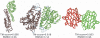

References
-
- Douguet D, Chen HC, Tovchigrechko A, Vakser IA. DOCKGROUND resource for studying protein-protein interfaces. Bioinformatics. 2006;22:2612–2618. - PubMed
-
- Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem. Sci. 1998;23:358–361. - PubMed
-
- Arakaki AK, Zhang Y, Skolnick J. Large-scale assessment of the utility of low-resolution protein structures for biochemical function assignment. Bioinformatics. 2004;20:1087–1096. - PubMed
-
- Graille M, Baltaze JP, Leulliot N, Liger D, Quevillon-Cheruel S, van Tilbeurgh H. Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase. J. Biol. Chem. 2006;281:30175–30185. - PubMed

