ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion
- PMID: 19443451
- PMCID: PMC2715244
- DOI: 10.1093/nar/gkp390
ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion
Abstract
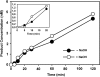
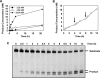
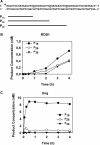
Arabidopsis ROS1 belongs to a family of plant 5-methycytosine DNA glycosylases that initiate DNA demethylation through base excision. ROS1 displays the remarkable capacity to excise 5-meC, and to a lesser extent T, while retaining the ability to discriminate effectively against C and U. We found that replacement of the C5-methyl group by halogen substituents greatly decreased excision of the target base. Furthermore, 5-meC was excised more efficiently from mismatches, whereas excision of T only occurred when mispaired with G. These results suggest that ROS1 specificity arises by a combination of selective recognition at the active site and thermodynamic stability of the target base. We also found that ROS1 is a low-turnover catalyst because it binds tightly to the abasic site left after 5-meC removal. This binding leads to a highly distributive behaviour of the enzyme on DNA substrates containing multiple 5-meC residues, and may help to avoid generation of double-strand breaks during processing of bimethylated CG dinucleotides. We conclude that the biochemical properties of ROS1 are consistent with its proposed role in protecting the plant genome from excess methylation.
Figures




References
-
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. - PubMed
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. - PubMed
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

