Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast
- PMID: 19443688
- PMCID: PMC2690032
- DOI: 10.1073/pnas.0813063106
Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast
Abstract
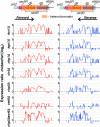
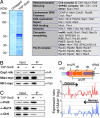
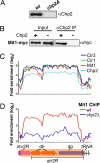
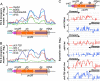
Conserved chromosomal HP1 proteins capable of binding to histone H3 methylated at lysine 9 are believed to provide a dynamic platform for the recruitment and/or spreading of various regulatory proteins involved in diverse chromosomal processes. The fission yeast Schizosaccharomyces pombe HP1 family members Chp2 and Swi6 are important for heterochromatin assembly and transcriptional silencing, but their precise roles are not fully understood. Here, we show that Swi6 and Chp2 associate with histone deacetylase (HDAC) protein complexes containing class I HDAC Clr6 and class II HDAC Clr3 (a component of Snf2/HDAC repressor complex), which are critical for transcriptional silencing of centromeric repeats targeted by the heterochromatin machinery. Mapping of RNA polymerase (Pol) II distribution in single and double mutant backgrounds revealed that Swi6 and Chp2 proteins and their associated HDAC complexes have overlapping functions in limiting Pol II occupancy across pericentromeric heterochromatin domains. The purified Swi6 fraction also contains factors involved in various chromosomal processes such as chromatin remodeling and DNA replication. Also, Swi6 copurifies with Mis4 protein, a cohesin loading factor essential for sister chromatid cohesion, and with centromere-specific histone H3 variant CENP-A, which is incorporated into chromatin in a heterochromatin-dependent manner. These analyses suggest that among other functions, HP1 proteins associate with chromatin-modifying factors that in turn cooperate to assemble repressive chromatin; thus, precluding accessibility of underlying DNA sequences to transcriptional machinery.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. - PubMed
-
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell. 2002;108:489–500. - PubMed
-
- Selker EU. Repeat-induced gene silencing in fungi. Adv Genet. 2002;46:439–450. - PubMed
-
- Birchler JA, Bhadra MP, Bhadra U. Making noise about silence: Repression of repeated genes in animals. Curr Opin Genet Dev. 2000;10:211–216. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

