Synthesis of orthogonal transcription-translation networks
- PMID: 19443689
- PMCID: PMC2689021
- DOI: 10.1073/pnas.0900267106
Synthesis of orthogonal transcription-translation networks
Abstract
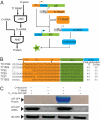
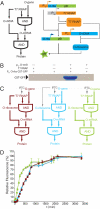
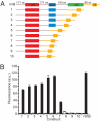
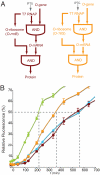
Orthogonal, parallel and independent, systems are one key foundation for synthetic biology. The synthesis of orthogonal systems that are uncoupled from evolutionary constraints, and selectively abstracted from cellular regulation, is an emerging approach to making biology more amenable to engineering. Here, we combine orthogonal transcription by T7 RNA polymerase and translation by orthogonal ribosomes (O-ribosomes), creating an orthogonal gene expression pathway in Escherichia coli. We design and implement compact, orthogonal gene expression networks. In particular we focus on creating transcription-translation feed-forward loops (FFLs). The transcription-translation FFLs reported cannot be created by using the cells' gene expression machinery and introduce information-processing delays on the order of hours into gene expression. We refactor the rRNA operon, uncoupling the synthesis of the orthogonal 16S rRNA for the O-ribosome from the synthesis and processing of the rest of the rRNA operon, thereby defining a minimal module that can be added to the cell for O-ribosome production. The minimal O-ribosome permits the rational alteration of the delay in an orthogonal gene expression FFL. Overall this work demonstrates that system-level dynamic properties are amenable to rational manipulation and design in orthogonal systems. In the future this system may be further evolved and tuned to provide a spectrum of tailored dynamics in gene expression and investigate the effects of delays in cellular decision-making processes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Chin JW. Modular approaches to expanding the functions of living matter. Nat Chem Biol. 2006;2:304–311. - PubMed
-
- Sprinzak D, Elowitz MB. Reconstruction of genetic circuits. Nature. 2005;438:443–448. - PubMed
-
- Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–554. - PubMed
-
- Kohanski MA, Collins JJ. Rewiring bacteria, two components at a time. Cell. 2008;133:947–948. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

