Toll-like receptors in control of immunological autophagy
- PMID: 19444282
- PMCID: PMC2788936
- DOI: 10.1038/cdd.2009.40
Toll-like receptors in control of immunological autophagy
Abstract
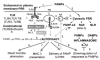
Autophagy is a cell biological process, enabling cells to autodigest their own cytosol when starved, remove cytoplasmic protein aggregates too large for proteasomal degradation, eliminate aberrant or over-proliferated organelles, and sanitize the cytoplasm by killing intracellular microbes. The role of autophagy has been expanded in recent years to include diverse immunological effector and regulatory functions. In this review, we summarize the multiple immunological roles of autophagy uncovered to date and focus primarily on details of induction of autophagy by pattern recognition receptors, as a newly established Toll-like receptor output. Taken together with other links between autophagy and innate and adaptive immunity processes, this cell-autonomous antimicrobial defense may be evolutionarily positioned at the root of immunity with the multiple innate and adaptive immunity connections uncovered to date reflecting a co-evolution of this ancient cell-defense mechanism and more advanced immunological systems in metazoans.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

