The Role of the NF-kappaB Transcriptome and Proteome as Biomarkers in Human Head and Neck Squamous Cell Carcinomas
- PMID: 19444329
- PMCID: PMC2681266
- DOI: 10.2217/17520363.2.4.409
The Role of the NF-kappaB Transcriptome and Proteome as Biomarkers in Human Head and Neck Squamous Cell Carcinomas
Abstract
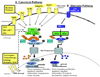
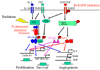
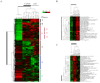
NF-kappaB is a family of signal activated transcription factors comprised of hetero- or homo-dimers from 5 different subunits, NF-kappaB1, NF-kappaB2, RELA, cREL and RELB. NF-kappaBs normally are transiently activated in response to infection or injury, but in cancers are aberrantly activated, regulating a transcriptome of hundreds of genes and corresponding proteome that promote pathogenesis and therapeutic resistance. In head and neck squamous cell carcinomas, an important role of NF-kappaB in regulation of the altered transcriptome and proteome has been established, providing a catalog of activating and target genes and proteins that may be useful as biomarkers of alterations in this pathway for this and other cancers. An emerging appreciation that NF-kappaB and other signal pathways form an altered regulatory network highlights the need to use biomarkers and combine targeted agents for personalized therapy of cancer.
Figures

Similar articles
-
Bortezomib-induced apoptosis with limited clinical response is accompanied by inhibition of canonical but not alternative nuclear factor-{kappa}B subunits in head and neck cancer.Clin Cancer Res. 2008 Jul 1;14(13):4175-85. doi: 10.1158/1078-0432.CCR-07-4470. Clin Cancer Res. 2008. PMID: 18593997
-
Lymphotoxin-β receptor-NIK signaling induces alternative RELB/NF-κB2 activation to promote metastatic gene expression and cell migration in head and neck cancer.Mol Carcinog. 2019 Mar;58(3):411-425. doi: 10.1002/mc.22938. Epub 2018 Nov 28. Mol Carcinog. 2019. PMID: 30488488 Free PMC article.
-
The RelA/cRel nuclear factor-κB (NF-κB) dimer, crucial for inflammation resolution, mediates the transcription of the key enzyme in melatonin synthesis in RAW 264.7 macrophages.J Pineal Res. 2016 May;60(4):394-404. doi: 10.1111/jpi.12321. Epub 2016 Mar 2. J Pineal Res. 2016. PMID: 26887983
-
Functions of NF-kappaB1 and NF-kappaB2 in immune cell biology.Biochem J. 2004 Sep 1;382(Pt 2):393-409. doi: 10.1042/BJ20040544. Biochem J. 2004. PMID: 15214841 Free PMC article. Review.
-
Nuclear factor-kappaB1: regulation and function.Int J Biochem Cell Biol. 2008;40(8):1425-30. doi: 10.1016/j.biocel.2007.05.004. Epub 2007 May 17. Int J Biochem Cell Biol. 2008. PMID: 17693123 Review.
Cited by
-
Gene expression profile alone is inadequate in predicting complete response in multiple myeloma.Leukemia. 2014 Nov;28(11):2229-34. doi: 10.1038/leu.2014.140. Epub 2014 Apr 15. Leukemia. 2014. PMID: 24732597 Free PMC article.
-
Circular RNA profiling reveals a potential role of hsa_circ_IPCEF1 in papillary thyroid carcinoma.Mol Med Rep. 2021 Aug;24(2):603. doi: 10.3892/mmr.2021.12241. Epub 2021 Jun 24. Mol Med Rep. 2021. PMID: 34165176 Free PMC article.
-
Head and neck cancer: pathogenesis and targeted therapy.MedComm (2020). 2024 Aug 21;5(9):e702. doi: 10.1002/mco2.702. eCollection 2024 Sep. MedComm (2020). 2024. PMID: 39170944 Free PMC article. Review.
-
Levels of circulating soluble receptor activator of NF-κB and interleukins-1 predicting outcome of locally advanced basal cell carcinoma.Int J Immunopathol Pharmacol. 2016 Dec;29(4):784-789. doi: 10.1177/0394632016675180. Epub 2016 Oct 19. Int J Immunopathol Pharmacol. 2016. PMID: 27760847 Free PMC article.
-
Curcumin Analogue CA15 Exhibits Anticancer Effects on HEp-2 Cells via Targeting NF-κB.Biomed Res Int. 2017;2017:4751260. doi: 10.1155/2017/4751260. Epub 2017 Mar 20. Biomed Res Int. 2017. PMID: 28409156 Free PMC article.
References
-
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. - PubMed
-
-
Van Waes C. Nuclear factor-kappaB in development, prevention, and therapy of cancer. Clin Cancer Res. 2007;13:1076–1082. **A brief but comprehensive review of evidence for role of NF-κB in development and as an investigational target for therapeutic and preventive agents
-
-
-
Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. **A comprehensive review of evidence for role of NF-κB in development of cancer
-
-
-
Basseres DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25:6817–6830. ** A comprehensive review of evidence for role of NF-κB in development of cancer
-
-
- Allen CT, Ricker JL, Chen Z, Van Waes C. Role of activated nuclear factor-kappaB in the pathogenesis and therapy of squamous cell carcinoma of the head and neck. Head Neck. 2007;29:959–971. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
