CHARMM: the biomolecular simulation program
- PMID: 19444816
- PMCID: PMC2810661
- DOI: 10.1002/jcc.21287
CHARMM: the biomolecular simulation program
Abstract
CHARMM (Chemistry at HARvard Molecular Mechanics) is a highly versatile and widely used molecular simulation program. It has been developed over the last three decades with a primary focus on molecules of biological interest, including proteins, peptides, lipids, nucleic acids, carbohydrates, and small molecule ligands, as they occur in solution, crystals, and membrane environments. For the study of such systems, the program provides a large suite of computational tools that include numerous conformational and path sampling methods, free energy estimators, molecular minimization, dynamics, and analysis techniques, and model-building capabilities. The CHARMM program is applicable to problems involving a much broader class of many-particle systems. Calculations with CHARMM can be performed using a number of different energy functions and models, from mixed quantum mechanical-molecular mechanical force fields, to all-atom classical potential energy functions with explicit solvent and various boundary conditions, to implicit solvent and membrane models. The program has been ported to numerous platforms in both serial and parallel architectures. This article provides an overview of the program as it exists today with an emphasis on developments since the publication of the original CHARMM article in 1983.
Copyright 2009 Wiley Periodicals, Inc.
Figures
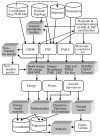

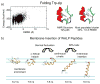




References
-
- Alder BJ, Wainwright TE. J Chem Phys. 1957;27:1208.
-
- Rahman A. Phys Rev. 1964;136:405–406.
-
- Rahman A, Stillinger FH. J Chem Phys. 1971;55:3336–3359.
-
- McCammon JA, Gelin BR, Karplus M. Nature. 1977;267:585–590. - PubMed
-
- Hockney RW, Eastwood JW. Computer Simulation Using Particles. McGraw-Hill; New York: 1981.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

