Analysis of four-way junctions in RNA structures
- PMID: 19445952
- PMCID: PMC2777522
- DOI: 10.1016/j.jmb.2009.04.084
Analysis of four-way junctions in RNA structures
Abstract
RNA secondary structures can be divided into helical regions composed of canonical Watson-Crick and related base pairs, as well as single-stranded regions such as hairpin loops, internal loops, and junctions. These elements function as building blocks in the design of diverse RNA molecules with various fundamental functions in the cell. To better understand the intricate architecture of three-dimensional (3D) RNAs, we analyze existing RNA four-way junctions in terms of base-pair interactions and 3D configurations. Specifically, we identify nine broad junction families according to coaxial stacking patterns and helical configurations. We find that helices within junctions tend to arrange in roughly parallel and perpendicular patterns and stabilize their conformations using common tertiary motifs such as coaxial stacking, loop-helix interaction, and helix packing interaction. Our analysis also reveals a number of highly conserved base-pair interaction patterns and novel tertiary motifs such as A-minor-coaxial stacking combinations and sarcin/ricin motif variants. Such analyses of RNA building blocks can ultimately help in the difficult task of RNA 3D structure prediction.
Figures

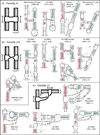


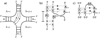
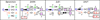

References
-
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. - PubMed
-
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. - PubMed
-
- Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. - PubMed
-
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. - PMC - PubMed
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

