Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex
- PMID: 19446455
- PMCID: PMC2788334
- DOI: 10.1016/j.cub.2009.04.058
Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex
Abstract
Background: Candida species are microbial pathogens originally thought to be asexual, but several are now recognized as sexual or parasexual. Candida albicans, the most common fungus infecting humans, is an obligate diploid with a parasexual cycle involving mating, recombination, and genome reduction but no recognized meiosis. Others (C. lusitaniae, C. guilliermondii) are haploid, and their mating produces spores, suggestive of complete meiotic sexual cycles. However, comparative genomic analysis reveals that these species lack key meiotic components, including the recombinase Dmc1 and cofactors (Mei5/Sae3), synaptonemal-complex proteins (Zip1-Zip4/Hop1), and the crossover interference pathway (Msh4/5).
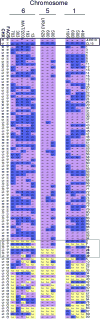
Results: Here we elucidate the structure and functions of the mating-type (MAT) locus and establish that C. lusitaniae undergoes meiosis during its sexual cycle. The MAT-encoded a2 (high-mobility group) and alpha1 (alpha domain) factors specify a and alpha cell identity, whereas the a1 homeodomain protein drives meiosis and sporulation and functions without its canonical heterodimeric partner, alpha2. Despite the apparent loss of meiotic genes, C. lusitaniae undergoes meiosis during sexual reproduction involving diploid intermediates, frequent SPO11-dependent recombination, and whole-genome reduction generating haploid progeny. The majority of meiotic progeny are euploid, but approximately one-third are diploid/aneuploid.
Conclusions: The cell identity and meiotic pathways have been substantially rewired, and meiotic generation of both recombinant and aneuploid progeny may expand genetic diversity. These findings inform our understanding of sexual reproduction in pathogenic microbes and the evolutionary plasticity of the meiotic machinery, with implications for the sexual nature of C. albicans and the generation and consequences of aneuploidy in biology and medicine.
Figures


Comment in
-
Sex: deviant mating in yeast.Curr Biol. 2009 Jul 14;19(13):R509-11. doi: 10.1016/j.cub.2009.05.026. Curr Biol. 2009. PMID: 19602410
References
-
- Hull CM, Johnson AD. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science. 1999;285:1271–1275. - PubMed
-
- Hull CM, Raisner RM, Johnson AD. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science. 2000;289:307–310. - PubMed
-
- Johnson A. The biology of mating in Candida albicans. Nat Rev Microbiol. 2003;1:106–116. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

