Data structures and compression algorithms for genomic sequence data
- PMID: 19447783
- PMCID: PMC2705231
- DOI: 10.1093/bioinformatics/btp319
Data structures and compression algorithms for genomic sequence data
Abstract
Motivation: The continuing exponential accumulation of full genome data, including full diploid human genomes, creates new challenges not only for understanding genomic structure, function and evolution, but also for the storage, navigation and privacy of genomic data. Here, we develop data structures and algorithms for the efficient storage of genomic and other sequence data that may also facilitate querying and protecting the data.
Results: The general idea is to encode only the differences between a genome sequence and a reference sequence, using absolute or relative coordinates for the location of the differences. These locations and the corresponding differential variants can be encoded into binary strings using various entropy coding methods, from fixed codes such as Golomb and Elias codes, to variables codes, such as Huffman codes. We demonstrate the approach and various tradeoffs using highly variables human mitochondrial genome sequences as a testbed. With only a partial level of optimization, 3615 genome sequences occupying 56 MB in GenBank are compressed down to only 167 KB, achieving a 345-fold compression rate, using the revised Cambridge Reference Sequence as the reference sequence. Using the consensus sequence as the reference sequence, the data can be stored using only 133 KB, corresponding to a 433-fold level of compression, roughly a 23% improvement. Extensions to nuclear genomes and high-throughput sequencing data are discussed.
Availability: Data are publicly available from GenBank, the HapMap web site, and the MITOMAP database. Supplementary materials with additional results, statistics, and software implementations are available from http://mammag.web.uci.edu/bin/view/Mitowiki/ProjectDNACompression.
Figures
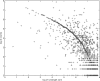
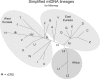
References
-
- Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Andrews R, et al. Reanalysis and revision of the cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;2:147. - PubMed
-
- Behzadi B, Fessant F. DNA compression challenge revisited: a dynamic programming approach. Lect. Notes Comput. Sci. 2005;3537:190–200.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

