Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance
- PMID: 19447905
- PMCID: PMC2704729
- DOI: 10.1128/JB.00144-09
Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance
Abstract
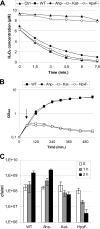
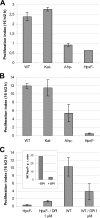
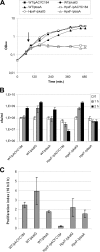
Salmonella enterica serovar Typhimurium is an intracellular pathogen that can survive and replicate within macrophages. One of the host defense mechanisms that Salmonella encounters during infection is the production of reactive oxygen species by the phagocyte NADPH oxidase. Among them, hydrogen peroxide (H(2)O(2)) can diffuse across bacterial membranes and damage biomolecules. Genome analysis allowed us to identify five genes encoding H(2)O(2) degrading enzymes: three catalases (KatE, KatG, and KatN) and two alkyl hydroperoxide reductases (AhpC and TsaA). Inactivation of the five cognate structural genes yielded the HpxF(-) mutant, which exhibited a high sensitivity to exogenous H(2)O(2) and a severe survival defect within macrophages. When the phagocyte NADPH oxidase was inhibited, its proliferation index increased 3.7-fold. Moreover, the overexpression of katG or tsaA in the HpxF(-) background was sufficient to confer a proliferation index similar to that of the wild type in macrophages and a resistance to millimolar H(2)O(2) in rich medium. The HpxF(-) mutant also showed an attenuated virulence in a mouse model. These data indicate that Salmonella catalases and alkyl hydroperoxide reductases are required to degrade H(2)O(2) and contribute to the virulence. This enzymatic redundancy highlights the evolutionary strategies developed by bacterial pathogens to survive within hostile environments.
Figures



Similar articles
-
The ABC-type efflux pump MacAB protects Salmonella enterica serovar typhimurium from oxidative stress.mBio. 2013 Oct 29;4(6):e00630-13. doi: 10.1128/mBio.00630-13. mBio. 2013. PMID: 24169575 Free PMC article.
-
Identification of the Vibrio vulnificus ahpCl gene and its influence on survival under oxidative stress and virulence.J Microbiol. 2009 Oct;47(5):624-32. doi: 10.1007/s12275-009-0130-x. Epub 2009 Oct 24. J Microbiol. 2009. PMID: 19851736
-
Cyclopropane Fatty Acids Are Important for Salmonella enterica Serovar Typhimurium Virulence.Infect Immun. 2022 Jan 25;90(1):e0047921. doi: 10.1128/IAI.00479-21. Epub 2021 Oct 18. Infect Immun. 2022. PMID: 34662213 Free PMC article.
-
Responses to reactive oxygen intermediates and virulence of Salmonella typhimurium.Microbes Infect. 2003 May;5(6):527-34. doi: 10.1016/s1286-4579(03)00069-8. Microbes Infect. 2003. PMID: 12758282 Review.
-
Peroxiredoxins and sports: new insights on the antioxidative defense.J Physiol Sci. 2013 Jan;63(1):1-5. doi: 10.1007/s12576-012-0237-4. Epub 2012 Oct 10. J Physiol Sci. 2013. PMID: 23055024 Free PMC article. Review.
Cited by
-
Salmonella Mitigates Oxidative Stress and Thrives in the Inflamed Gut by Evading Calprotectin-Mediated Manganese Sequestration.Cell Host Microbe. 2016 Jun 8;19(6):814-25. doi: 10.1016/j.chom.2016.05.005. Cell Host Microbe. 2016. PMID: 27281571 Free PMC article.
-
Metabolic Feedback Inhibition Influences Metabolite Secretion by the Human Gut Symbiont Bacteroides thetaiotaomicron.mSystems. 2020 Sep 1;5(5):e00252-20. doi: 10.1128/mSystems.00252-20. mSystems. 2020. PMID: 32873608 Free PMC article.
-
Genetic regulation of the ompX porin of Salmonella Typhimurium in response to hydrogen peroxide stress.Biol Res. 2022 Feb 22;55(1):8. doi: 10.1186/s40659-022-00377-3. Biol Res. 2022. PMID: 35193678 Free PMC article.
-
Bacterial Membrane-Derived Vesicles Attenuate Vancomycin Activity against Methicillin-Resistant Staphylococcus aureus.Microorganisms. 2021 Sep 29;9(10):2055. doi: 10.3390/microorganisms9102055. Microorganisms. 2021. PMID: 34683376 Free PMC article.
-
Functional Characterization of Salmonella Typhimurium Encoded YciF, a Domain of Unknown Function (DUF892) Family Protein, and Its Role in Protection during Bile and Oxidative Stress.J Bacteriol. 2023 Jul 25;205(7):e0005923. doi: 10.1128/jb.00059-23. Epub 2023 Jun 27. J Bacteriol. 2023. PMID: 37367303 Free PMC article.
References
-
- Babior, B. M. 1999. NADPH oxidase: an update. Blood 931464-1476. - PubMed
-
- Beuzón, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 31345-1352. - PubMed
-
- Bryk, R., P. Griffin, and C. Nathan. 2000. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407211-215. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

