Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis
- PMID: 19447910
- PMCID: PMC2704730
- DOI: 10.1128/JB.00315-09
Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis
Abstract
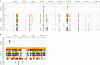
Nosocomial infections caused by antibiotic-resistant Klebsiella pneumoniae are emerging as a major health problem worldwide, while community-acquired K. pneumoniae infections present with a range of diverse clinical pictures in different geographic areas. In particular, an invasive form of K. pneumoniae that causes liver abscesses was first observed in Asia and then was found worldwide. We are interested in how differences in gene content of the same species result in different diseases. Thus, we sequenced the whole genome of K. pneumoniae NTUH-K2044, which was isolated from a patient with liver abscess and meningitis, and analyzed differences compared to strain MGH 78578, which was isolated from a patient with pneumonia. Six major types of differences were found in gene clusters that included an integrative and conjugative element, clusters involved in citrate fermentation, lipopolysaccharide synthesis, and capsular polysaccharide synthesis, phage-related insertions, and a cluster containing fimbria-related genes. We also conducted comparative genomic hybridization with 15 K. pneumoniae isolates obtained from community-acquired or nosocomial infections using tiling probes for the NTUH-K2044 genome. Hierarchical clustering revealed three major groups of genomic insertion-deletion patterns that correlate with the strains' clinical features, antimicrobial susceptibilities, and virulence phenotypes with mice. Here we report the whole-genome sequence of K. pneumoniae NTUH-K2044 and describe evidence showing significant genomic diversity and sequence acquisition among K. pneumoniae pathogenic strains. Our findings support the hypothesis that these factors are responsible for the changes that have occurred in the disease profile over time.
Figures


References
-
- Balows, A., and B. Duerden. 1998. Topley & Wilson's microbiology and microbial infections, 9th ed., vol. 2. Oxford University Press, Oxford, United Kingdom.
-
- Borodovsky, M., and J. McIninch. 1993. GeneMark: parallel gene recognition for both DNA strands. Comput. Chem. 17123-133.
-
- Brisse, S., and J. Verhoef. 2001. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int. J. Syst. Evol. Microbiol. 51915-924. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

