A screen for modifiers of notch signaling uncovers Amun, a protein with a critical role in sensory organ development
- PMID: 19448274
- PMCID: PMC2728848
- DOI: 10.1534/genetics.108.099986
A screen for modifiers of notch signaling uncovers Amun, a protein with a critical role in sensory organ development
Abstract
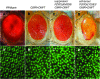
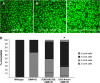
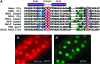
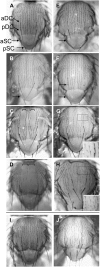
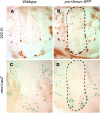
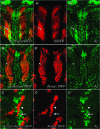
Notch signaling is an evolutionarily conserved pathway essential for many cell fate specification events during metazoan development. We conducted a large-scale transposon-based screen in the developing Drosophila eye to identify genes involved in Notch signaling. We screened 10,447 transposon lines from the Exelixis collection for modifiers of cell fate alterations caused by overexpression of the Notch ligand Delta and identified 170 distinct modifier lines that may affect up to 274 genes. These include genes known to function in Notch signaling, as well as a large group of characterized and uncharacterized genes that have not been implicated in Notch pathway function. We further analyze a gene that we have named Amun and show that it encodes a protein that localizes to the nucleus and contains a putative DNA glycosylase domain. Genetic and molecular analyses of Amun show that altered levels of Amun function interfere with cell fate specification during eye and sensory organ development. Overexpression of Amun decreases expression of the proneural transcription factor Achaete, and sensory organ loss caused by Amun overexpression can be rescued by coexpression of Achaete. Taken together, our data suggest that Amun acts as a transcriptional regulator that can affect cell fate specification by controlling Achaete levels.
Figures


Similar articles
-
Identification of 11-amino acid peptides that disrupt Notch-mediated processes in Drosophila.J Biomed Sci. 2011 Jun 17;18(1):42. doi: 10.1186/1423-0127-18-42. J Biomed Sci. 2011. PMID: 21682860 Free PMC article.
-
The Drosophila T-box transcription factor Midline functions within the Notch-Delta signaling pathway to specify sensory organ precursor cell fates and regulates cell survival within the eye imaginal disc.Mech Dev. 2013 Nov-Dec;130(11-12):577-601. doi: 10.1016/j.mod.2013.08.001. Epub 2013 Aug 17. Mech Dev. 2013. PMID: 23962751 Free PMC article.
-
Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye.Curr Biol. 2001 Mar 6;11(5):330-8. doi: 10.1016/s0960-9822(01)00093-8. Curr Biol. 2001. PMID: 11267869
-
Robust selection of sensory organ precursors by the Notch-Delta pathway.Curr Opin Cell Biol. 2011 Dec;23(6):663-7. doi: 10.1016/j.ceb.2011.09.005. Epub 2011 Sep 29. Curr Opin Cell Biol. 2011. PMID: 21963301 Review.
-
Hairless: the ignored antagonist of the Notch signalling pathway.Hereditas. 2006 Dec;143(2006):212-21. doi: 10.1111/j.2007.0018-0661.01971.x. Hereditas. 2006. PMID: 17362357 Review.
Cited by
-
Genome-wide association analysis of oxidative stress resistance in Drosophila melanogaster.PLoS One. 2012;7(4):e34745. doi: 10.1371/journal.pone.0034745. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496853 Free PMC article.
-
The Drosophila junctophilin gene is functionally equivalent to its four mammalian counterparts and is a modifier of a Huntingtin poly-Q expansion and the Notch pathway.Dis Model Mech. 2018 Jan 17;11(1):dmm029082. doi: 10.1242/dmm.029082. Dis Model Mech. 2018. PMID: 29208631 Free PMC article.
-
Tools and methods for studying Notch signaling in Drosophila melanogaster.Methods. 2014 Jun 15;68(1):173-82. doi: 10.1016/j.ymeth.2014.03.029. Epub 2014 Apr 3. Methods. 2014. PMID: 24704358 Free PMC article.
-
Regulation of Drosophila intestinal stem cell maintenance and differentiation by the transcription factor Escargot.EMBO J. 2014 Dec 17;33(24):2983-96. doi: 10.15252/embj.201489050. Epub 2014 Nov 27. EMBO J. 2014. PMID: 25433031 Free PMC article.
-
Integration of Drosophila and Human Genetics to Understand Notch Signaling Related Diseases.Adv Exp Med Biol. 2018;1066:141-185. doi: 10.1007/978-3-319-89512-3_8. Adv Exp Med Biol. 2018. PMID: 30030826 Free PMC article. Review.
References
-
- Artavanis-Tsakonas, S., 2004. Accessing the Exelixis collection. Nat. Genet. 36 207. - PubMed
-
- Artavanis-Tsakonas, S., M. D. Rand and R. J. Lake, 1999. Notch signalling: cell fate control and signal integration in development. Science 284 770–775. - PubMed
-
- Baker, K. D., L. M. Shewchuk, T. Kozlova, M. Makishima, A. Hassell et al., 2003. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell 113 731–742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

