c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry
- PMID: 19448666
- PMCID: PMC2779836
- DOI: 10.1038/onc.2009.112
c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry
Abstract
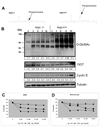
Cell proliferation requires the coordinated activity of cytosolic and mitochondrial metabolic pathways to provide ATP and building blocks for DNA, RNA and protein synthesis. Many metabolic pathway genes are targets of the c-myc oncogene and cell-cycle regulator. However, the contribution of c-Myc to the activation of cytosolic and mitochondrial metabolic networks during cell-cycle entry is unknown. Here, we report the metabolic fates of [U-(13)C] glucose in serum-stimulated myc(-/-) and myc(+/+) fibroblasts by (13)C isotopomer NMR analysis. We demonstrate that endogenous c-myc increased (13)C labeling of ribose sugars, purines and amino acids, indicating partitioning of glucose carbons into C1/folate and pentose phosphate pathways, and increased tricarboxylic acid cycle turnover at the expense of anaplerotic flux. Myc expression also increased global O-linked N-acetylglucosamine protein modification, and inhibition of hexosamine biosynthesis selectively reduced growth of Myc-expressing cells, suggesting its importance in Myc-induced proliferation. These data reveal a central organizing function for the Myc oncogene in the metabolism of cycling cells. The pervasive deregulation of this oncogene in human cancers may be explained by its function in directing metabolic networks required for cell proliferation.
Figures


Similar articles
-
The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry.Cell Cycle. 2008 Apr 15;7(8):1054-66. doi: 10.4161/cc.7.8.5739. Epub 2008 Feb 8. Cell Cycle. 2008. PMID: 18414044 Free PMC article.
-
c-Myc programs fatty acid metabolism and dictates acetyl-CoA abundance and fate.J Biol Chem. 2014 Sep 5;289(36):25382-92. doi: 10.1074/jbc.M114.580662. Epub 2014 Jul 22. J Biol Chem. 2014. PMID: 25053415 Free PMC article.
-
Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry.J Biol Chem. 2010 Nov 19;285(47):36267-74. doi: 10.1074/jbc.M110.141606. Epub 2010 Sep 2. J Biol Chem. 2010. PMID: 20813845 Free PMC article.
-
MYC, Metabolism, and Cancer.Cancer Discov. 2015 Oct;5(10):1024-39. doi: 10.1158/2159-8290.CD-15-0507. Epub 2015 Sep 17. Cancer Discov. 2015. PMID: 26382145 Free PMC article. Review.
-
MYC and metabolism on the path to cancer.Semin Cell Dev Biol. 2015 Jul;43:11-21. doi: 10.1016/j.semcdb.2015.08.003. Epub 2015 Aug 12. Semin Cell Dev Biol. 2015. PMID: 26277543 Free PMC article. Review.
Cited by
-
tPA is a potent mitogen for renal interstitial fibroblasts: role of beta1 integrin/focal adhesion kinase signaling.Am J Pathol. 2010 Sep;177(3):1164-75. doi: 10.2353/ajpath.2010.091269. Epub 2010 Jul 16. Am J Pathol. 2010. PMID: 20639453 Free PMC article.
-
c-MYC and HIF1α promoter G-quadruplexes dependent metabolic regulation mechanism of berberine in colon cancer.J Gastrointest Oncol. 2022 Jun;13(3):1152-1168. doi: 10.21037/jgo-22-389. J Gastrointest Oncol. 2022. PMID: 35837174 Free PMC article.
-
Metabolic Effects of Recurrent Genetic Aberrations in Multiple Myeloma.Cancers (Basel). 2021 Jan 21;13(3):396. doi: 10.3390/cancers13030396. Cancers (Basel). 2021. PMID: 33494394 Free PMC article. Review.
-
The many lives of Myc in the pancreatic β-cell.J Biol Chem. 2021 Jan-Jun;296:100122. doi: 10.1074/jbc.REV120.011149. Epub 2020 Dec 2. J Biol Chem. 2021. PMID: 33239359 Free PMC article. Review.
-
Rethinking the Warburg effect with Myc micromanaging glutamine metabolism.Cancer Res. 2010 Feb 1;70(3):859-62. doi: 10.1158/0008-5472.CAN-09-3556. Epub 2010 Jan 19. Cancer Res. 2010. PMID: 20086171 Free PMC article. Review.
References
-
- Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. - PubMed
-
- Dean M, Levine RA, Ran W, Kindy MS, Sonenshein GE, Campisi J. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J Biol Chem. 1986;261:9161–9166. - PubMed
-
- Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. - PubMed
-
- Hunt AN, Postle AD. Phosphatidylcholine biosynthesis inside the nucleus: is it involved in regulating cell proliferation? Adv Enzyme Regul. 2004;44:173–186. - PubMed
-
- Jackowski S. Coordination of membrane phospholipid synthesis with the cell cycle. J Biol Chem. 1994;269:3858–3867. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

