HIV latency: present knowledge, future directions
- PMID: 19448843
- PMCID: PMC2682531
- DOI: 10.2217/17460794.1.6.733
HIV latency: present knowledge, future directions
Abstract
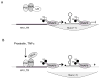
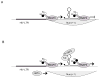
Current therapies do not eradicate HIV from infected patients. Indeed, HIV hides in a latent form insensitive to these therapies. Thus, one priority is to purge these latent reservoirs. But what mechanisms are responsible for latency and what are the reservoirs of latently infected cells? The present knowledge in terms of HIV latency is still incomplete and current therapeutic strategies fail to eradicate completely latently infected cells. What could the future bring?
Figures




References
-
- Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1(12):1284–90. The first study in which stable form of latent HIV was found in purified resting CD4+ T-lymphocytes. - PubMed
-
- Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–7. Demonstrates the importance of latent infection of resting CD4+ T cells for viral persistence and predicts the long time required for eradication of the latent reservoir. - PubMed
-
- Takahashi K, Wesselingh SL, Griffin DE, McArthur JC, Johnson RT, Glass JD. Localization of HIV-1 in human brain using polymerase chain reaction/in situ hybridization and immunocytochemistry. Ann Neurol. 1996;39(6):705–11. - PubMed
-
- Geijtenbeek TB, Torensma R, van Vliet SJ, et al. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100(5):575–85. - PubMed
-
- Igarashi T, Imamichi H, Brown CR, Hirsch VM, Martin MA. The emergence and characterization of macrophage-tropic SIV/HIV chimeric viruses (SHIVs) present in CD4+ T cell-depleted rhesus monkeys. J Leukoc Biol. 2003;74(5):772–80. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
