Nicotinamide/nicotinic acid mononucleotide adenylyltransferase, new insights into an ancient enzyme
- PMID: 19448972
- PMCID: PMC11115848
- DOI: 10.1007/s00018-009-0047-x
Nicotinamide/nicotinic acid mononucleotide adenylyltransferase, new insights into an ancient enzyme
Abstract
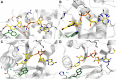
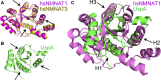
Nicotinamide/nicotinic acid mononucleotide adenylyltransferase (NMNAT) has long been known as the master enzyme in NAD biosynthesis in living organisms. A burst of investigations on NMNAT, going beyond enzymology, have paralleled increasing discoveries of key roles played by NAD homeostasis in a number or patho-physiological conditions. The availability of in-depth kinetics and structural enzymology analyses carried out on NMNATs from different organisms offer a powerful tool for uncovering fascinating evolutionary relationships. On the other hand, additional functions featuring NMNAT have emerged from investigations aimed at unraveling the molecular mechanisms responsible for complex biological phenomena such as neurodegeneration. NMNAT appears to be a multifunctional protein that sits both at the core of central metabolism and at a crossroads of multiple cellular processes. The resultant wealth of biochemical data has built a robust framework upon which design of NMNAT activators, inhibitors or enzyme variants of potential medical interest can be based.
Figures




References
-
- Gerdes SY, Scholle MD, D’Souza M, Bernal A, Baev MV, Farrell M, Kurnasov OV, Daugherty MD, Mseeh F, Polanuyer BM, Campbell JW, Anantha S, Shatalin KY, Chowdhury SA, Fonstein MY, Osterman AL. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J Bacteriol. 2002;184:4555–4572. doi: 10.1128/JB.184.16.4555-4572.2002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

