Single nucleotide polymorphism genotyping in polyploid wheat with the Illumina GoldenGate assay
- PMID: 19449174
- PMCID: PMC2715469
- DOI: 10.1007/s00122-009-1059-5
Single nucleotide polymorphism genotyping in polyploid wheat with the Illumina GoldenGate assay
Abstract
Single nucleotide polymorphisms (SNPs) are indispensable in such applications as association mapping and construction of high-density genetic maps. These applications usually require genotyping of thousands of SNPs in a large number of individuals. Although a number of SNP genotyping assays are available, most of them are designed for SNP genotyping in diploid individuals. Here, we demonstrate that the Illumina GoldenGate assay could be used for SNP genotyping of homozygous tetraploid and hexaploid wheat lines. Genotyping reactions could be carried out directly on genomic DNA without the necessity of preliminary PCR amplification. A total of 53 tetraploid and 38 hexaploid homozygous wheat lines were genotyped at 96 SNP loci. The genotyping error rate estimated after removal of low-quality data was 0 and 1% for tetraploid and hexaploid wheat, respectively. Developed SNP genotyping assays were shown to be useful for genotyping wheat cultivars. This study demonstrated that the GoldenGate assay is a very efficient tool for high-throughput genotyping of polyploid wheat, opening new possibilities for the analysis of genetic variation in wheat and dissection of genetic basis of complex traits using association mapping approach.
Figures
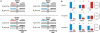

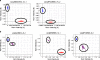
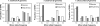
References
-
- Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang SY, Uszynski G, Mohler V, Lehmensiek A, Kuchel H, Hayden MJ, Howes N, Sharp P, Vaughan P, Rathmell B, Huttner E, Kilian A (2006) Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet 113:1409–1420 - DOI - PubMed
-
- Akhunov ED, Goodyear AW, Geng S, Qi LL, Echalier B, Gill BS, Miftahudin Gustafson JP, Lazo G, Chao SM, Anderson OD, Linkiewicz AM, Dubcovsky J, La Rota M, Sorrells ME, Zhang DS, Nguyen HT, Kalavacharla V, Hossain K, Kianian SF, Peng JH, Lapitan NLV, Gonzalez-Hernandeiz JL, Anderson JA, Choi DW, Close TJ, Dilbirligi M, Gill KS, Walker-Simmons MK, Steber C, McGuire PE, Qualset CO, Dvorak J (2003) The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res 13:753–763 - DOI - PMC - PubMed
-
- Aranzana MJ, Kim S, Zhao K, Bakker E, Horton M, Jakob K, Lister C, Molitor J, Shindo C, Tang C, Toomajian C, Traw B, Zheng H, Bergelson J, Dean C, Marjoram P, Nordborg M (2005) Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet 1:e60 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

