Regulation of expression of the Haemophilus ducreyi LspB and LspA2 proteins by CpxR
- PMID: 19451237
- PMCID: PMC2715676
- DOI: 10.1128/IAI.00292-09
Regulation of expression of the Haemophilus ducreyi LspB and LspA2 proteins by CpxR
Abstract
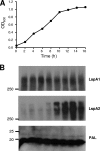
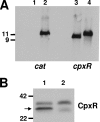
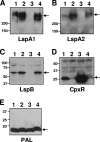
The LspA1, LspA2, and LspB proteins of Haemophilus ducreyi comprise a two-partner secretion system that has been shown to be necessary for H. ducreyi to inhibit phagocytosis by immune cells in vitro. Inactivation of lspA1 resulted in increased levels of LspA2, suggesting that these two proteins are differentially controlled (C. J. Ward et al., Infect. Immun. 71:2478-2486, 2003). Expression of LspA2 but not LspA1 was shown to be both growth phase dependent and affected by the presence of fetal calf serum (FCS) in the growth medium. In addition, neither LspA1 nor LspA2 could be detected in culture supernatant fluid in the absence of FCS. DNA microarray analysis revealed that 324 H. ducreyi genes were differentially regulated after growth in the presence of FCS. Among these, the CpxRA two-component sensory transduction system was downregulated by the presence of FCS. Inactivation of cpxR resulted in increased expression of both LspB and LspA2. Electrophoretic mobility shift assays showed that a recombinant H. ducreyi CpxR protein bound the promoter region of the lspB-lspA2 operon. The cpxR and cpxA genes were shown to be part of an operon containing two additional genes in H. ducreyi 35000HP. This is the first description of a two-component sensory transduction system regulating a proven virulence factor of H. ducreyi.
Figures

References
-
- Albritton, W. L., I. W. MacLean, P. D. Bertram, and A. R. Ronald. 1981. Haemin requirements in Haemophilus with special reference to H. ducreyi, p. 75-82. In M. Kilian, W. Frederiksen, and E. L. Biberstein (ed.), Haemophilus, Pasteurella, and Actinobacillus. Academic Press, London, England.
-
- Al Tawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 1811049-1054. - PubMed
-
- Anonymous. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

