An immunogenic, surface-exposed domain of Haemophilus ducreyi outer membrane protein HgbA is involved in hemoglobin binding
- PMID: 19451245
- PMCID: PMC2708535
- DOI: 10.1128/IAI.00034-09
An immunogenic, surface-exposed domain of Haemophilus ducreyi outer membrane protein HgbA is involved in hemoglobin binding
Abstract
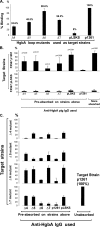
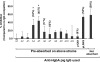
HgbA is the sole TonB-dependent receptor for hemoglobin (Hb) acquisition of Haemophilus ducreyi. Binding of Hb to HgbA is the initial step in heme acquisition from Hb. To better understand this step, we mutagenized hgbA by deletion of each of the 11 putative surface-exposed loops and expressed each of the mutant proteins in trans in host strain H. ducreyi FX547 hgbA. All mutant proteins were expressed, exported, and detected on the surface by anti-HgbA immunoglobulin G (IgG). Deletion of sequences in loops 5 and 7 of HgbA abolished Hb binding in two different formats. In contrast, HgbA proteins containing deletions in the other nine loops retained the ability to bind Hb. None of the clones expressing mutant proteins were able to grow on plates containing low concentrations of Hb. Previously we demonstrated in a swine model of chancroid infection that an HgbA vaccine conferred complete protection from a challenge infection. Using anti-HgbA IgG from this study and the above deletion mutants, we show that loops 4, 5, and 7 of HgbA were immunogenic and surface exposed and that IgG directed against loops 4 and 5 blocked Hb binding. Furthermore, loop 6 was cleaved by protease on intact H. ducreyi, suggesting surface exposure. These data implicate a central domain of HgbA (in respect to the primary amino acid sequence) as important in Hb binding and suggest that this region of the molecule might have potential as a subunit vaccine.
Figures



Similar articles
-
Role of the Haemophilus ducreyi Ton system in internalization of heme from hemoglobin.Infect Immun. 1998 Jan;66(1):151-60. doi: 10.1128/IAI.66.1.151-160.1998. Infect Immun. 1998. PMID: 9423852 Free PMC article.
-
Immunization with the Haemophilus ducreyi hemoglobin receptor HgbA protects against infection in the swine model of chancroid.Infect Immun. 2006 Apr;74(4):2224-32. doi: 10.1128/IAI.74.4.2224-2232.2006. Infect Immun. 2006. PMID: 16552053 Free PMC article.
-
Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi.Infect Immun. 1995 Apr;63(4):1241-5. doi: 10.1128/iai.63.4.1241-1245.1995. Infect Immun. 1995. PMID: 7890379 Free PMC article.
-
Mutational analysis of hemoglobin binding and heme utilization by a bacterial hemoglobin receptor.J Bacteriol. 2013 Jul;195(13):3115-23. doi: 10.1128/JB.00199-13. Epub 2013 May 10. J Bacteriol. 2013. PMID: 23667232 Free PMC article.
-
Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi.Infect Immun. 1995 Jun;63(6):2194-200. doi: 10.1128/iai.63.6.2194-2200.1995. Infect Immun. 1995. PMID: 7768598 Free PMC article.
Cited by
-
Heme uptake in bacterial pathogens.Curr Opin Chem Biol. 2014 Apr;19:34-41. doi: 10.1016/j.cbpa.2013.12.014. Epub 2014 Jan 4. Curr Opin Chem Biol. 2014. PMID: 24780277 Free PMC article. Review.
-
Host-pathogen interplay of Haemophilus ducreyi.Curr Opin Infect Dis. 2010 Feb;23(1):64-9. doi: 10.1097/QCO.0b013e328334c0cb. Curr Opin Infect Dis. 2010. PMID: 19918177 Free PMC article. Review.
-
Dissecting binding of a β-barrel membrane protein by phage display.Mol Biosyst. 2017 Jul 25;13(8):1438-1447. doi: 10.1039/c7mb00163k. Mol Biosyst. 2017. PMID: 28627567 Free PMC article.
-
Extracellular heme uptake and the challenges of bacterial cell membranes.Curr Top Membr. 2012;69:359-92. doi: 10.1016/B978-0-12-394390-3.00013-6. Curr Top Membr. 2012. PMID: 23046657 Free PMC article.
-
Haemophilus ducreyi Infection Induces Oxidative Stress, Central Metabolic Changes, and a Mixed Pro- and Anti-inflammatory Environment in the Human Host.mBio. 2022 Dec 20;13(6):e0312522. doi: 10.1128/mbio.03125-22. Epub 2022 Dec 1. mBio. 2022. PMID: 36453940 Free PMC article.
References
-
- Albritton, W. L., I. W. Macklean, P. D. Bertram, and A. R. Ronald. 1981. Haemin requirements in Haemophilus with special reference to H. ducreyi. Academic Press, Inc., New York, NY.
-
- Al-Tawfiq, J. A., A. C. Thornton, B. P. Katz, K. R. Fortney, K. D. Todd, A. F. Hood, and S. M. Spinola. 1998. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J. Infect. Dis. 1781684-1687. - PubMed
-
- Al-Tawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 1811049-1054. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

