Heterochromatin protein 1 is recruited to various types of DNA damage
- PMID: 19451271
- PMCID: PMC2711568
- DOI: 10.1083/jcb.200810035
Heterochromatin protein 1 is recruited to various types of DNA damage
Abstract
Heterochromatin protein 1 (HP1) family members are chromatin-associated proteins involved in transcription, replication, and chromatin organization. We show that HP1 isoforms HP1-alpha, HP1-beta, and HP1-gamma are recruited to ultraviolet (UV)-induced DNA damage and double-strand breaks (DSBs) in human cells. This response to DNA damage requires the chromo shadow domain of HP1 and is independent of H3K9 trimethylation and proteins that detect UV damage and DSBs. Loss of HP1 results in high sensitivity to UV light and ionizing radiation in the nematode Caenorhabditis elegans, indicating that HP1 proteins are essential components of DNA damage response (DDR) systems. Analysis of single and double HP1 mutants in nematodes suggests that HP1 homologues have both unique and overlapping functions in the DDR. Our results show that HP1 proteins are important for DNA repair and may function to reorganize chromatin in response to damage.
Figures
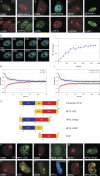
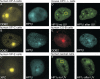
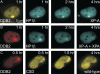
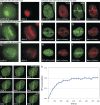
Comment in
-
Revisiting the role of heterochromatin protein 1 in DNA repair.J Cell Biol. 2009 May 18;185(4):573-5. doi: 10.1083/jcb.200904033. J Cell Biol. 2009. PMID: 19451270 Free PMC article. Review.
References
-
- Agarwal B.K., Sparrow J.H. 1981. Line intensities in the soft x-ray region.J. Phys. F Met. Phys. 11:1303–1309
-
- Aten J.A., Stap J., Krawczyk P.M., van Oven C.H., Hoebe R.A., Essers J., Kanaar R. 2004. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains.Science. 303:92–95 - PubMed
-
- Ayoub N., Jeyasekharan A.D., Bernal J.A., Venkitaraman A.R. 2008. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response.Nature. 453:682–686 - PubMed
-
- Bartek J., Lukas J. 2007. DNA damage checkpoints: from initiation to recovery or adaptation.Curr. Opin. Cell Biol. 19:238–245 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

