DExD/H-box Prp5 protein is in the spliceosome during most of the splicing cycle
- PMID: 19451545
- PMCID: PMC2704087
- DOI: 10.1261/rna.1065209
DExD/H-box Prp5 protein is in the spliceosome during most of the splicing cycle
Abstract
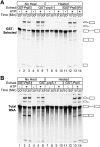
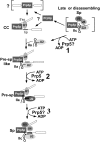
The DExD/H-box Prp5 protein (Prp5p) is an essential, RNA-dependent ATPase required for pre-spliceosome formation during nuclear pre-mRNA splicing. In order to understand how this protein functions, we used in vitro, biochemical assays to examine its association with the spliceosome from Saccharomyces cerevisiae. GST-Prp5p in splicing assays pulls down radiolabeled pre-mRNA as well as splicing intermediates and lariat product, but reduced amounts of spliced mRNA. It cosediments with active spliceosomes isolated by glycerol gradient centrifugation. In ATP-depleted extracts, GST-Prp5p associates with pre-mRNA even in the absence of spliceosomal snRNAs. Maximal selection in either the presence or absence of ATP requires a pre-mRNA with a functional intron. Prp5p is present in the commitment complex and functions in subsequent pre-spliceosome formation. Reduced Prp5p levels decrease levels of commitment, pre-spliceosomal and spliceosomal complexes. Thus Prp5p is most likely an integral component of the spliceosome, being among the first splicing factors associating with pre-mRNA and remaining until spliceosome disassembly. The results suggest a model in which Prp5p recruits the U2 snRNP to pre-mRNA in the commitment complex and then hydrolyzes ATP to promote stable association of U2 in the pre-spliceosome. They also suggest that Prp5p could have multiple ATP-independent and ATP-dependent functions at several stages of the splicing cycle.
Figures






References
-
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. - PubMed
-
- Abu Dayyeh B, Quan TK, Castro MA, Ruby SW. Probing interactions between the U2 snRNP and the DEAD-box protein, Prp5. J Biol Chem. 2002;277:20221–20233. - PubMed
-
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. - PubMed
-
- Brody E, Abelson J. The “spliceosome”: Yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985;228:963–967. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
