Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis
- PMID: 19451621
- PMCID: PMC2683883
- DOI: 10.1073/pnas.0812937106
Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis
Abstract
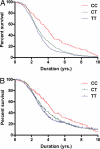
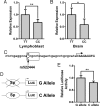
Amyotrophic lateral sclerosis is a degenerative disorder of motor neurons that typically develops in the 6th decade and is uniformly fatal, usually within 5 years. To identify genetic variants associated with susceptibility and phenotypes in sporadic ALS, we performed a genome-wide SNP analysis in sporadic ALS cases and controls. A total of 288,357 SNPs were screened in a set of 1,821 sporadic ALS cases and 2,258 controls from the U.S. and Europe. Survival analysis was performed using 1,014 deceased sporadic cases. Top results for susceptibility were further screened in an independent sample set of 538 ALS cases and 556 controls. SNP rs1541160 within the KIFAP3 gene (encoding a kinesin-associated protein) yielded a genome-wide significant result (P = 1.84 x 10(-8)) that withstood Bonferroni correction for association with survival. Homozygosity for the favorable allele (CC) conferred a 14.0 months survival advantage. Sequence, genotypic and functional analyses revealed that there is linkage disequilibrium between rs1541160 and SNP rs522444 within the KIFAP3 promoter and that the favorable alleles of rs1541160 and rs522444 correlate with reduced KIFAP3 expression. No SNPs were associated with risk of sporadic ALS, site of onset, or age of onset. We have identified a variant within the KIFAP3 gene that is associated with decreased KIFAP3 expression and increased survival in sporadic ALS. These findings support the view that genetic factors modify phenotypes in this disease and that cellular motor proteins are determinants of motor neuron viability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Tandan R, Bradley WG. Amyotrophic lateral sclerosis: Part 1. Clinical features, pathology and ethical issues in management. Ann Neurol. 1985;18:271–280. - PubMed
-
- Tandan R, Bradley WG. Amyotrophic lateral sclerosis: Part 2. Etiopathogenesis. Ann Neurol. 1985;18:419–431. - PubMed
-
- Mulder D, Kurland L, Offord K, Beard C. Familial adult motor neuron disease: Amyotrophic lateral sclerosis. Neurology. 1986;36:511–517. - PubMed
-
- Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

