Farnesylation of pex19p is required for its structural integrity and function in peroxisome biogenesis
- PMID: 19451657
- PMCID: PMC2742854
- DOI: 10.1074/jbc.M109.016584
Farnesylation of pex19p is required for its structural integrity and function in peroxisome biogenesis
Abstract
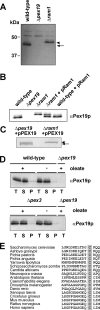
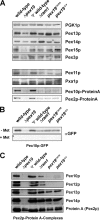
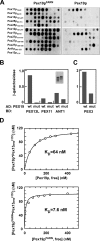
The conserved CaaX box peroxin Pex19p is known to be modified by farnesylation. The possible involvement of this lipid modification in peroxisome biogenesis, the degree to which Pex19p is farnesylated, and its molecular function are unknown or controversial. We resolve these issues by first showing that the complete pool of Pex19p is processed by farnesyltransferase in vivo and that this modification is independent of peroxisome induction or the Pex19p membrane anchor Pex3p. Furthermore, genomic mutations of PEX19 prove that farnesylation is essential for proper matrix protein import into peroxisomes, which is supposed to be caused indirectly by a defect in peroxisomal membrane protein (PMP) targeting or stability. This assumption is corroborated by the observation that mutants defective in Pex19p farnesylation are characterized by a significantly reduced steady-state concentration of prominent PMPs (Pex11p, Ant1p) but also of essential components of the peroxisomal import machinery, especially the RING peroxins, which were almost depleted from the importomer. In vivo and in vitro, PMP recognition is only efficient when Pex19p is farnesylated with affinities differing by a factor of 10 between the non-modified and wild-type forms of Pex19p. Farnesylation is likely to induce a conformational change in Pex19p. Thus, isoprenylation of Pex19p contributes to substrate membrane protein recognition for the topogenesis of PMPs, and our results highlight the importance of lipid modifications in protein-protein interactions.
Figures



Similar articles
-
Farnesylation of Pex19p is not essential for peroxisome biogenesis in yeast and mammalian cells.Cell Mol Life Sci. 2006 Jul;63(14):1686-99. doi: 10.1007/s00018-006-6110-y. Cell Mol Life Sci. 2006. PMID: 16791427 Free PMC article.
-
Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions.Biochim Biophys Acta. 2006 Dec;1763(12):1639-46. doi: 10.1016/j.bbamcr.2006.09.030. Epub 2006 Sep 26. Biochim Biophys Acta. 2006. PMID: 17069900 Review.
-
Pex19p binds Pex30p and Pex32p at regions required for their peroxisomal localization but separate from their peroxisomal targeting signals.J Biol Chem. 2006 May 26;281(21):14805-12. doi: 10.1074/jbc.M601808200. Epub 2006 Mar 21. J Biol Chem. 2006. PMID: 16551610
-
Functional domain mapping of peroxin Pex19p: interaction with Pex3p is essential for function and translocation.J Cell Sci. 2006 Sep 1;119(Pt 17):3539-50. doi: 10.1242/jcs.03100. Epub 2006 Aug 8. J Cell Sci. 2006. PMID: 16895967
-
Peroxisome biogenesis disorders: molecular basis for impaired peroxisomal membrane assembly: in metabolic functions and biogenesis of peroxisomes in health and disease.Biochim Biophys Acta. 2012 Sep;1822(9):1337-42. doi: 10.1016/j.bbadis.2012.06.004. Epub 2012 Jun 13. Biochim Biophys Acta. 2012. PMID: 22705440 Review.
Cited by
-
Involvement of the carboxyl-terminal region of the yeast peroxisomal half ABC transporter Pxa2p in its interaction with Pxa1p and in transporter function.PLoS One. 2014 Aug 13;9(8):e104892. doi: 10.1371/journal.pone.0104892. eCollection 2014. PLoS One. 2014. PMID: 25118695 Free PMC article.
-
Peroxisome biogenesis, membrane contact sites, and quality control.EMBO Rep. 2019 Jan;20(1):e46864. doi: 10.15252/embr.201846864. Epub 2018 Dec 10. EMBO Rep. 2019. PMID: 30530632 Free PMC article. Review.
-
Selective isotope labeling for NMR structure determination of proteins in complex with unlabeled ligands.J Biomol NMR. 2019 Apr;73(3-4):183-189. doi: 10.1007/s10858-019-00241-9. Epub 2019 Apr 30. J Biomol NMR. 2019. PMID: 31041647 Free PMC article.
-
Engineering a membrane protein chaperone to ameliorate the proteotoxicity of mutant huntingtin.Nat Commun. 2025 Jan 17;16(1):737. doi: 10.1038/s41467-025-56030-6. Nat Commun. 2025. PMID: 39824813 Free PMC article.
-
Functional regions of the peroxin Pex19 necessary for peroxisome biogenesis.J Biol Chem. 2017 Jul 7;292(27):11547-11560. doi: 10.1074/jbc.M116.774067. Epub 2017 May 19. J Biol Chem. 2017. PMID: 28526747 Free PMC article.
References
-
- Young S. G., Fong L. G., Michaelis S. (2005) J. Lipid Res. 46, 2531–2558 - PubMed
-
- Sebti S. M., Der C. J. (2003) Nat. Rev. Cancer 3, 945–951 - PubMed
-
- Zhang F. L., Casey P. J. (1996) Annu. Rev. Biochem. 65, 241–269 - PubMed
-
- Magee T., Seabra M. C. (2005) Curr. Opin. Cell Biol. 17, 190–196 - PubMed
-
- Sinensky M. (2000) Biochim. Biophys. Acta 1529, 203–209 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

