Episcopic 3D Imaging Methods: Tools for Researching Gene Function
- PMID: 19452045
- PMCID: PMC2682936
- DOI: 10.2174/138920208784533601
Episcopic 3D Imaging Methods: Tools for Researching Gene Function
Abstract
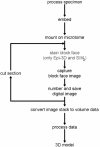
This work aims at describing episcopic 3D imaging methods and at discussing how these methods can contribute to researching the genetic mechanisms driving embryogenesis and tissue remodelling, and the genesis of pathologies. Several episcopic 3D imaging methods exist. The most advanced are capable of generating high-resolution volume data (voxel sizes from 0.5x0.5x1 microm upwards) of small to large embryos of model organisms and tissue samples. Beside anatomy and tissue architecture, gene expression and gene product patterns can be three dimensionally analyzed in their precise anatomical and histological context with the aid of whole mount in situ hybridization or whole mount immunohistochemical staining techniques. Episcopic 3D imaging techniques were and are employed for analyzing the precise morphological phenotype of experimentally malformed, randomly produced, or genetically engineered embryos of biomedical model organisms. It has been shown that episcopic 3D imaging also fits for describing the spatial distribution of genes and gene products during embryogenesis, and that it can be used for analyzing tissue samples of adult model animals and humans. The latter offers the possibility to use episcopic 3D imaging techniques for researching the causality and treatment of pathologies or for staging cancer. Such applications, however, are not yet routine and currently only preliminary results are available. We conclude that, although episcopic 3D imaging is in its very beginnings, it represents an upcoming methodology, which in short terms will become an indispensable tool for researching the genetic regulation of embryo development as well as the genesis of malformations and diseases.
Keywords: 3D modelling; development; embryo; episcopic microscopy; gene expression.; imaging.
Figures


Similar articles
-
Visualising the Cardiovascular System of Embryos of Biomedical Model Organisms with High Resolution Episcopic Microscopy (HREM).J Cardiovasc Dev Dis. 2018 Dec 15;5(4):58. doi: 10.3390/jcdd5040058. J Cardiovasc Dev Dis. 2018. PMID: 30558275 Free PMC article. Review.
-
Three-dimensional analysis of molecular signals with episcopic imaging techniques.Methods Mol Biol. 2007;411:35-46. doi: 10.1007/978-1-59745-549-7_4. Methods Mol Biol. 2007. PMID: 18287637
-
High-resolution episcopic microscopy: a rapid technique for high detailed 3D analysis of gene activity in the context of tissue architecture and morphology.Anat Embryol (Berl). 2006 Jun;211(3):213-21. doi: 10.1007/s00429-005-0073-x. Epub 2006 Jan 21. Anat Embryol (Berl). 2006. PMID: 16429276
-
Visualizing vertebrate embryos with episcopic 3D imaging techniques.ScientificWorldJournal. 2009 Dec 16;9:1423-37. doi: 10.1100/tsw.2009.154. ScientificWorldJournal. 2009. PMID: 20024516 Free PMC article. Review.
-
microMRI-HREM pipeline for high-throughput, high-resolution phenotyping of murine embryos.J Anat. 2007 Jul;211(1):132-7. doi: 10.1111/j.1469-7580.2007.00746.x. Epub 2007 May 28. J Anat. 2007. PMID: 17532797 Free PMC article.
Cited by
-
Three-dimensional structural and metric characterisation of cardioids.Front Cell Dev Biol. 2024 Jul 25;12:1426043. doi: 10.3389/fcell.2024.1426043. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39119041 Free PMC article.
-
High-Resolution Episcopic Microscopy (HREM) in Multimodal Imaging Approaches.Biomedicines. 2021 Dec 15;9(12):1918. doi: 10.3390/biomedicines9121918. Biomedicines. 2021. PMID: 34944735 Free PMC article. Review.
-
3-dimensional imaging modalities for phenotyping genetically engineered mice.Vet Pathol. 2012 Jan;49(1):106-15. doi: 10.1177/0300985811429814. Epub 2011 Dec 6. Vet Pathol. 2012. PMID: 22146851 Free PMC article. Review.
-
Spatial Change of Cruciate Ligaments in Rat Embryo Knee Joint by Three-Dimensional Reconstruction.PLoS One. 2015 Jun 22;10(6):e0131092. doi: 10.1371/journal.pone.0131092. eCollection 2015. PLoS One. 2015. PMID: 26098761 Free PMC article.
-
Phase-contrast X-ray microtomography of mouse fetus.Biol Open. 2012 Mar 15;1(3):269-74. doi: 10.1242/bio.2012430. Epub 2012 Feb 10. Biol Open. 2012. PMID: 23213417 Free PMC article.
References
-
- Seltmann M, Horsch M, Drobyshev A, Chen Y, de Angelis MH, Beckers J. Assessment of a systematic expression profiling approach in ENU-induced mouse mutant lines. Mamm. Genome. 2005;16:1–10. - PubMed
-
- Reecy JM, Spurlock DM, Stahl CH. Gene expression profiling: insights into skeletal muscle growth and development. J. Anim. Sci. 2006;84(Suppl):E150–154. - PubMed
-
- Beckers J, Hoheisel J, Mewes W, Vingron M, Hrabe de Angelis MH. Molecular phenotyping of mouse mutant resources by RNA expression profiling. Curr. Genomics. 2002;3:121–129.
-
- Mitiku N, Baker JC. Genomic analysis of gastrulation and organogenesis in the mouse. Dev. Cell. 2007;13:897–907. - PubMed
LinkOut - more resources
Full Text Sources
