Engineered cystine knot peptides that bind alphavbeta3, alphavbeta5, and alpha5beta1 integrins with low-nanomolar affinity
- PMID: 19452550
- PMCID: PMC5792193
- DOI: 10.1002/prot.22441
Engineered cystine knot peptides that bind alphavbeta3, alphavbeta5, and alpha5beta1 integrins with low-nanomolar affinity
Abstract
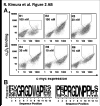
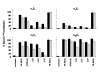
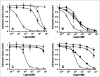
There is a critical need for compounds that target cell surface integrin receptors for applications in cancer therapy and diagnosis. We used directed evolution to engineer the Ecballium elaterium trypsin inhibitor (EETI-II), a knottin peptide from the squash family of protease inhibitors, as a new class of integrin-binding agents. We generated yeast-displayed libraries of EETI-II by substituting its 6-amino acid trypsin binding loop with 11-amino acid loops containing the Arg-Gly-Asp integrin binding motif and randomized flanking residues. These libraries were screened in a high-throughput manner by fluorescence-activated cell sorting to identify mutants that bound to alpha(v)beta(3) integrin. Select peptides were synthesized and were shown to compete for natural ligand binding to integrin receptors expressed on the surface of U87MG glioblastoma cells with half-maximal inhibitory concentration values of 10-30 nM. Receptor specificity assays demonstrated that engineered knottin peptides bind to both alpha(v)beta(3) and alpha(v)beta(5) integrins with high affinity. Interestingly, we also discovered a peptide that binds with high affinity to alpha(v)beta(3), alpha(v)beta(5), and alpha(5)beta(1) integrins. This finding has important clinical implications because all three of these receptors can be coexpressed on tumors. In addition, we showed that engineered knottin peptides inhibit tumor cell adhesion to the extracellular matrix protein vitronectin, and in some cases fibronectin, depending on their integrin binding specificity. Collectively, these data validate EETI-II as a scaffold for protein engineering, and highlight the development of unique integrin-binding peptides with potential for translational applications in cancer.
Figures

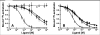
References
-
- Folkman J. The role of angiogenesis in tumor growth. Semin Cancer Biol. 1992;3:65–71. - PubMed
-
- Alghisi GC, Ruegg C. Vascular integrins in tumor angiogenesis: mediators and therapeutic targets. Endothelium. 2006;13:113–135. - PubMed
-
- Brooks PC, Clark RA, Cheresh DA. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science. 1994;264:569–571. - PubMed
-
- Mizejewski GJ. Role of integrins in cancer: survey of expression patterns. Proc Soc Exp Biol Med. 1999;222:124–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

