Proteomics of Porphyromonas gingivalis within a model oral microbial community
- PMID: 19454014
- PMCID: PMC2689231
- DOI: 10.1186/1471-2180-9-98
Proteomics of Porphyromonas gingivalis within a model oral microbial community
Abstract
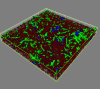
Background: Porphyromonas gingivalis is a periodontal pathogen that resides in a complex multispecies microbial biofilm community known as dental plaque. Confocal laser scanning microscopy showed that P. gingivalis can assemble into communities in vitro with Streptococcus gordonii and Fusobacterium nucleatum, common constituents of dental plaque. Whole cell quantitative proteomics, along with mutant construction and analysis, were conducted to investigate how P. gingivalis adapts to this three species community.
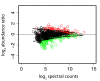
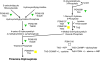
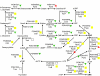
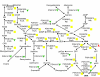
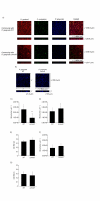
Results: 1156 P. gingivalis proteins were detected qualitatively during comparison of the three species model community with P. gingivalis incubated alone under the same conditions. Integration of spectral counting and summed signal intensity analyses of the dataset showed that 403 proteins were down-regulated and 89 proteins up-regulated. The proteomics results were inspected manually and an ontology analysis conducted using DAVID. Significant decreases were seen in proteins involved in cell shape and the formation of the cell envelope, as well as thiamine, cobalamin, and pyrimidine synthesis and DNA repair. An overall increase was seen in proteins involved in protein synthesis. HmuR, a TonB dependent outer membrane receptor, was up-regulated in the community and an hmuR deficient mutant was deficient in three species community formation, but was unimpaired in its ability to form mono- or dual-species biofilms.
Conclusion: Collectively, these results indicate that P. gingivalis can assemble into a heterotypic community with F. nucleatum and S. gordonii, and that a community lifestyle provides physiologic support for P. gingivalis. Proteins such as HmuR, that are up-regulated, can be necessary for community structure.
Figures
Similar articles
-
Proteomics of Streptococcus gordonii within a model developing oral microbial community.BMC Microbiol. 2012 Sep 18;12:211. doi: 10.1186/1471-2180-12-211. BMC Microbiol. 2012. PMID: 22989070 Free PMC article.
-
Proteomics of Fusobacterium nucleatum within a model developing oral microbial community.Microbiologyopen. 2014 Oct;3(5):729-51. doi: 10.1002/mbo3.204. Epub 2014 Aug 25. Microbiologyopen. 2014. PMID: 25155235 Free PMC article.
-
Quantitative proteomic analysis of extracellular matrix extracted from mono- and dual-species biofilms of Fusobacterium nucleatum and Porphyromonas gingivalis.Anaerobe. 2017 Apr;44:133-142. doi: 10.1016/j.anaerobe.2017.03.002. Epub 2017 Mar 9. Anaerobe. 2017. PMID: 28285095
-
Halitosis vaccines targeting FomA, a biofilm-bridging protein of fusobacteria nucleatum.Curr Mol Med. 2013 Sep;13(8):1358-67. doi: 10.2174/15665240113139990063. Curr Mol Med. 2013. PMID: 23865430 Review.
-
Spheres of influence: Porphyromonas gingivalis outer membrane vesicles.Mol Oral Microbiol. 2016 Oct;31(5):365-78. doi: 10.1111/omi.12134. Epub 2015 Oct 23. Mol Oral Microbiol. 2016. PMID: 26466922 Review.
Cited by
-
Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection.Nat Microbiol. 2017 Nov;2(11):1493-1499. doi: 10.1038/s41564-017-0021-6. Epub 2017 Sep 18. Nat Microbiol. 2017. PMID: 28924191 Free PMC article.
-
Proteomics of Streptococcus gordonii within a model developing oral microbial community.BMC Microbiol. 2012 Sep 18;12:211. doi: 10.1186/1471-2180-12-211. BMC Microbiol. 2012. PMID: 22989070 Free PMC article.
-
Arginine-Ornithine Antiporter ArcD Controls Arginine Metabolism and Interspecies Biofilm Development of Streptococcus gordonii.J Biol Chem. 2015 Aug 28;290(35):21185-98. doi: 10.1074/jbc.M115.644401. Epub 2015 Jun 17. J Biol Chem. 2015. PMID: 26085091 Free PMC article.
-
Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel.J Bacteriol. 2009 Nov;191(22):6804-11. doi: 10.1128/JB.01006-09. Epub 2009 Sep 11. J Bacteriol. 2009. PMID: 19749049 Free PMC article.
-
Intercellular communications in multispecies oral microbial communities.Front Microbiol. 2014 Jul 1;5:328. doi: 10.3389/fmicb.2014.00328. eCollection 2014. Front Microbiol. 2014. PMID: 25071741 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

