Conformational flexibility and molecular interactions of an archaeal homologue of the Shwachman-Bodian-Diamond syndrome protein
- PMID: 19454024
- PMCID: PMC2695463
- DOI: 10.1186/1472-6807-9-32
Conformational flexibility and molecular interactions of an archaeal homologue of the Shwachman-Bodian-Diamond syndrome protein
Abstract
Background: Defects in the human Shwachman-Bodian-Diamond syndrome (SBDS) protein-coding gene lead to the autosomal recessive disorder characterised by bone marrow dysfunction, exocrine pancreatic insufficiency and skeletal abnormalities. This protein is highly conserved in eukaryotes and archaea but is not found in bacteria. Although genomic and biophysical studies have suggested involvement of this protein in RNA metabolism and in ribosome biogenesis, its interacting partners remain largely unknown.
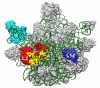
Results: We determined the crystal structure of the SBDS orthologue from Methanothermobacter thermautotrophicus (mthSBDS). This structure shows that SBDS proteins are highly flexible, with the N-terminal FYSH domain and the C-terminal ferredoxin-like domain capable of undergoing substantial rotational adjustments with respect to the central domain. Affinity chromatography identified several proteins from the large ribosomal subunit as possible interacting partners of mthSBDS. Moreover, SELEX (Systematic Evolution of Ligands by EXponential enrichment) experiments, combined with electrophoretic mobility shift assays (EMSA) suggest that mthSBDS does not interact with RNA molecules in a sequence specific manner.
Conclusion: It is suggested that functional interactions of SBDS proteins with their partners could be facilitated by rotational adjustments of the N-terminal and the C-terminal domains with respect to the central domain. Examination of the SBDS protein structure and domain movements together with its possible interaction with large ribosomal subunit proteins suggest that these proteins could participate in ribosome function.
Figures
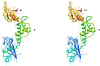
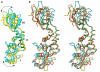
Similar articles
-
Structure, dynamics, and RNA interaction analysis of the human SBDS protein.J Mol Biol. 2010 Mar 5;396(4):1053-69. doi: 10.1016/j.jmb.2009.12.039. Epub 2010 Jan 4. J Mol Biol. 2010. PMID: 20053358
-
Structural and mutational analysis of the SBDS protein family. Insight into the leukemia-associated Shwachman-Diamond Syndrome.J Biol Chem. 2005 May 13;280(19):19221-9. doi: 10.1074/jbc.M414656200. Epub 2005 Feb 8. J Biol Chem. 2005. PMID: 15701631
-
Phylogeny, sequence conservation, and functional complementation of the SBDS protein family.Genomics. 2006 Jun;87(6):758-71. doi: 10.1016/j.ygeno.2006.01.010. Epub 2006 Mar 10. Genomics. 2006. PMID: 16529906
-
Clinical spectrum and molecular pathophysiology of Shwachman-Diamond syndrome.Curr Opin Hematol. 2011 Jan;18(1):30-5. doi: 10.1097/MOH.0b013e32834114a5. Curr Opin Hematol. 2011. PMID: 21124213 Free PMC article. Review.
-
Structure of a two-domain N-terminal fragment of ribosomal protein L10 from Methanococcus jannaschii reveals a specific piece of the archaeal ribosomal stalk.J Mol Biol. 2010 Jun 4;399(2):214-20. doi: 10.1016/j.jmb.2010.04.017. Epub 2010 Apr 24. J Mol Biol. 2010. PMID: 20399793 Review.
Cited by
-
Normal Mode Analysis as a Routine Part of a Structural Investigation.Molecules. 2019 Sep 10;24(18):3293. doi: 10.3390/molecules24183293. Molecules. 2019. PMID: 31510014 Free PMC article. Review.
-
Essential ribosome assembly factor Fap7 regulates a hierarchy of RNA-protein interactions during small ribosomal subunit biogenesis.Proc Natl Acad Sci U S A. 2013 Sep 17;110(38):15253-8. doi: 10.1073/pnas.1306389110. Epub 2013 Sep 3. Proc Natl Acad Sci U S A. 2013. PMID: 24003121 Free PMC article.
-
Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome.Adv Biol Regul. 2018 Jan;67:109-127. doi: 10.1016/j.jbior.2017.09.002. Epub 2017 Sep 6. Adv Biol Regul. 2018. PMID: 28942353 Free PMC article. Review.
-
Interaction of the GTPase Elongation Factor Like-1 with the Shwachman-Diamond Syndrome Protein and Its Missense Mutations.Int J Mol Sci. 2018 Dec 12;19(12):4012. doi: 10.3390/ijms19124012. Int J Mol Sci. 2018. PMID: 30545121 Free PMC article.
-
Non-Diamond Blackfan anemia disorders of ribosome function: Shwachman Diamond syndrome and 5q- syndrome.Semin Hematol. 2011 Apr;48(2):136-43. doi: 10.1053/j.seminhematol.2011.01.002. Semin Hematol. 2011. PMID: 21435510 Free PMC article. Review.
References
-
- Boocock GRB, Marit MR, Rommens JM. Phylogeny, sequence conservation, and functional complementation of the SBDS protein family. Genomics. 2006;87:758–771. - PubMed
-
- Erdos M, Alapi K, Balogh I, Oroszlan G, Rakoczi E, Sumegi J, Marodi L. Severe Shwachman-Diamond syndrome phenotype caused by compound heterozygous missense mutations in the SBDS gene. Exp Hematol. 2006;34:1517–1521. - PubMed
-
- Nakashima E, Mabuchi A, Makita Y, Masuno M, Ohashi H, Nishimura G, Ikegawa S. Novel SBDS mutations caused by gene conversion in Japanese patients with Shwachman-Diamond syndrome. Hum Genet. 2004;114:345–348. - PubMed
-
- Taneichi H, Kanegane H, Futatani T, Otsubo K, Nomura K, Sato Y, Hama A, Kojima S, Kohdera U, Nakano T, Hori H, Kawashima H, Inoh Y, Kamizono J, Adachi N, Osugi Y, Mizuno H, Hotta N, Yoneyama H, Nakashima E, Ikegawa S, Miyawaki T. Clinical and genetic analyses of presumed Shwachman-Diamond syndrome in Japan. Int J Hematol. 2006;84:60–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 077371/WT_/Wellcome Trust/United Kingdom
- BB/F001134/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F003099/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0500367/MRC_/Medical Research Council/United Kingdom
- BB/D525056/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

