Transcriptomic and proteomic analysis of global ischemia and cardioprotection in the rabbit heart
- PMID: 19454556
- PMCID: PMC2712218
- DOI: 10.1152/physiolgenomics.00033.2009
Transcriptomic and proteomic analysis of global ischemia and cardioprotection in the rabbit heart
Abstract
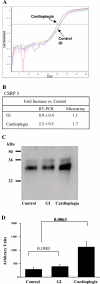
Cardioplegia is used to partially alleviate the effects of surgically induced global ischemia injury; however, the molecular mechanisms involved in this cardioprotection remain to be elucidated. To improve the understanding of the molecular processes modulating the effects of global ischemia and the cardioprotection afforded by cardioplegia, we constructed rabbit heart cDNA libraries and isolated, sequenced, and identified a compendium of nonredundant cDNAs for use in transcriptomic and proteomic analyses. New Zealand White rabbits were used to compare the effects of global ischemia and cardioplegia compared with control (nonischemic) hearts. The effects of RNA and protein synthesis on the cardioprotection afforded by cardioplegia were investigated separately by preperfusion with either alpha-amanitin or cycloheximide. Our results demonstrate that cardioplegia partially ameliorates the effects of global ischemia and that the cardioprotection is modulated by RNA- and protein-dependent mechanisms. Transcriptomic and proteomic enrichment analyses indicated that global ischemia downregulated genes/proteins associated with mitochondrial function and energy production, cofactor catabolism, and the generation of precursor metabolites of energy. In contrast, cardioplegia significantly increased differentially expressed genes/proteins associated with the mitochondrion and mitochondrial function and significantly upregulated the biological processes of muscle contraction, involuntary muscle contraction, carboxylic acid and fatty acid catabolic processes, fatty acid beta-oxidation, and fatty acid metabolic processes.
Figures




References
-
- Altman N Replication, variation and normalization in microarray experiments. Appl Bioinformatics 4: 33–44, 2005. - PubMed
-
- Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kääb S, Pfeufer A, Uberfuhr P, Dugas M, Steinbeck G, Nabauer M. Functional profiling of human atrial and ventricular gene expression. Pflügers Arch 50: 201–208, 2005. - PubMed
-
- Cardin S, Pelletier P, Libby E, Le Bouter S, Xiao L, Kääb S, Demolombe S, Glass L, Nattel S. Marked differences between atrial and ventricular gene-expression remodeling in dogs with experimental heart failure. J Mol Cell Cardiol 45: 821–831, 2008. - PubMed
-
- Crozier SJ, Bolster DR, Reiter AK, Kimball SR, Jefferson LS. β-Oxidation of free fatty acids is required to maintain translational control of protein synthesis in the heart. Am J Physiol Endocrinol Metab 283: E1144–E1150, 2002. - PubMed
-
- Eddy LJ, Stewart JR, Jones HP, Engerson TD, McCord JM, Downey JM. Free radical-producing enzyme, xanthine oxidase, is undetectable in human hearts. Am J Physiol Heart Circ Physiol 253: H709–H711, 1987. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

