Deciphering signaling outcomes from a system of complex networks
- PMID: 19454649
- PMCID: PMC2711542
- DOI: 10.1126/scisignal.2000054
Deciphering signaling outcomes from a system of complex networks
Abstract
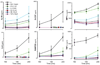
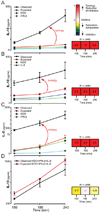
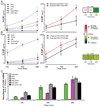
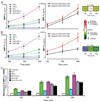
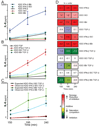
Cellular signal transduction machinery integrates information from multiple inputs to actuate discrete cellular behaviors. Interaction complexity exists when an input modulates the output behavior that results from other inputs. To address whether this machinery is iteratively complex--that is, whether increasing numbers of inputs produce exponential increases in discrete cellular behaviors--we examined the modulated secretion of six cytokines from macrophages in response to up to five-way combinations of an agonist of Toll-like receptor 4, three cytokines, and conditions that activated the cyclic adenosine monophosphate pathway. Although all of the selected ligands showed synergy in paired combinations, few examples of nonadditive outputs were found in response to higher-order combinations. This suggests that most potential interactions are not realized and that unique cellular responses are limited to discrete subsets of ligands and pathways that enhance specific cellular functions.
Figures




References
-
- Bagchi A, Herrup EA, Warren HS, Trigilio J, Shin H-S, Valentine C, Hellman J. MyD88-Dependent and MyD88-Independent Pathways in Synergy, Priming, and Tolerance between TLR Agonists. J Immunol. 2007;178:1164–1171. - PubMed
-
- Gouwy M, Struyf S, Proost P, Van Damme J. Synergy in cytokine and chemokine networks amplifies the inflammatory response. Cytokine Growth Factor Rev. 2005;16:561–580. - PubMed
-
- Natarajan M, Lin KM, Hsueh RC, Sternweis PC, Ranganathan R. A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol. 2006;8:571–580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

