Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB
- PMID: 19454733
- PMCID: PMC2700530
- DOI: 10.1105/tpc.108.062935
Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB
Abstract
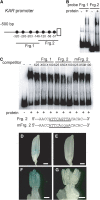
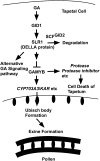
Gibberellins (GAs) play important roles in regulating reproductive development, especially anther development. Our previous studies revealed that the MYB transcriptional factor GAMYB, an important component of GA signaling in cereal aleurone cells, is also important for anther development. Here, we examined the physiological functions of GA during anther development through phenotypic analyses of rice (Oryza sativa) GA-deficient, GA-insensitive, and gamyb mutants. The mutants exhibited common defects in programmed cell death (PCD) of tapetal cells and formation of exine and Ubisch bodies. Microarray analysis using anther RNAs of these mutants revealed that rice GAMYB is involved in almost all instances of GA-regulated gene expression in anthers. Among the GA-regulated genes, we focused on two lipid metabolic genes, a cytochrome P450 hydroxylase CYP703A3 and beta-ketoacyl reductase, both of which might be involved in providing a substrate for exine and Ubisch body. GAMYB specifically interacted with GAMYB binding motifs in the promoter regions in vitro, and mutation of these motifs in promoter-beta-glucuronidase (GUS) transformants caused reduced GUS expression in anthers. Furthermore, a knockout mutant for CYP703A3 showed gamyb-like defects in exine and Ubisch body formation. Together, these results suggest that GA regulates exine formation and the PCD of tapetal cells and that direct activation of CYP703A3 by GAMYB is key to exine formation.
Figures








References
-
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131 3357–3365. - PubMed
-
- Ahlers, F., Bubert, H., Steuernagel, S., and Wiermann, R. (2000). The nature of oxygen in sporopollenin from the pollen of Typha angustifolia L. Z. Naturforsch. [C] 55 129–136. - PubMed
-
- Ahlers, F., Thom, I., Lambert, J., Kuckuk, R., and Wiermann, R. (1999). 1H NMR analysis of sporopollenin from Typha angustifolia. Phytochemistry 50 1095–1098.
-
- Bagnall, D.J. (1992). Control of flowering in Arabidopsis thaliana by light, vernalisation and gibberellins. Aust. J. Plant Physiol. 19 401–409.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

