Protein sequestration generates a flexible ultrasensitive response in a genetic network
- PMID: 19455136
- PMCID: PMC2694680
- DOI: 10.1038/msb.2009.30
Protein sequestration generates a flexible ultrasensitive response in a genetic network
Abstract
Ultrasensitive responses are crucial for cellular regulation. Protein sequestration, where an active protein is bound in an inactive complex by an inhibitor, can potentially generate ultrasensitivity. Here, in a synthetic genetic circuit in budding yeast, we show that sequestration of a basic leucine zipper transcription factor by a dominant-negative inhibitor converts a graded transcriptional response into a sharply ultrasensitive response, with apparent Hill coefficients up to 12. A simple quantitative model for this genetic network shows that both the threshold and the degree of ultrasensitivity depend upon the abundance of the inhibitor, exactly as we observed experimentally. The abundance of the inhibitor can be altered by simple mutation; thus, ultrasensitive responses mediated by protein sequestration are easily tuneable. Gene duplication of regulatory homodimers and loss-of-function mutations can create dominant negatives that sequester and inactivate the original regulator. The generation of flexible ultrasensitive responses is an unappreciated adaptive advantage that could explain the frequent evolutionary emergence of dominant negatives.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
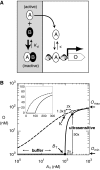
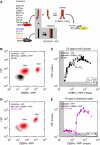
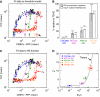
References
-
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H (1990) The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 61: 49–59 - PubMed
-
- Buchler NE, Louis M (2008) Molecular titration and ultrasensitivity in regulatory networks. J Mol Biol 384: 1106–1119 - PubMed
-
- Burke D, Dawson D, Stearns T (2000) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

