Structural basis for the sugar nucleotide and acyl-chain selectivity of Leptospira interrogans LpxA
- PMID: 19456129
- PMCID: PMC2710806
- DOI: 10.1021/bi900629e
Structural basis for the sugar nucleotide and acyl-chain selectivity of Leptospira interrogans LpxA
Abstract
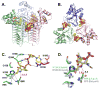
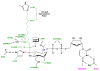
The first step of lipid A biosynthesis is catalyzed by LpxA in Escherichia coli (EcLpxA), an acyltransferase selective for UDP-GlcNAc and R-3-hydroxymyristoyl-acyl carrier protein (ACP). Leptospira interrogans LpxA (LiLpxA) is extremely selective for R-3-hydroxylauroyl-ACP and an analogue of UDP-GlcNAc, designated UDP-GlcNAc3N, in which NH(2) replaces the GlcNAc 3-OH group. EcLpxA does not discriminate between UDP-GlcNAc and UDP-GlcNAc3N; however, E. coli does not make UDP-GlcNAc3N. With LiLpxA, R-3-hydroxylauroyl-methylphosphopantetheine efficiently substitutes for R-3-hydroxylauroyl-ACP. We now present crystal structures of free LiLpxA and its complexes with its product UDP-3-N-(R-3-hydroxylauroyl)-GlcNAc3N and with its substrate R-3-hydroxylauroyl-methylphosphopantetheine. The positions of the acyl chains of the R-3-hydroxylauroyl-methylphosphopantetheine and the UDP-3-N-(R-3-hydroxylauroyl)-GlcNAc3N are almost identical and are similar to that of the acyl chain in the EcLpxA/UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc complex. The selectivity of LiLpxA for UDP-GlcNAc3N may be explained by the orientation of the backbone carbonyl group of Q68, which differs by approximately 82 degrees from the corresponding Q73 carbonyl group in EcLpxA. This arrangement provides an extra hydrogen-bond acceptor for the 3-NH(2) group of UDP-GlcNAc3N in LiLpxA. The R-3-hydroxylauroyl selectivity of LiLpxA is explained by the position of the K171 side chain, which limits the length of the acyl-chain-binding groove. Our results support the role of LiLpxA H120 (which corresponds to EcLpxA H125) as the catalytic base and provide the first structural information about the orientation of the phosphopantetheine moiety during LpxA catalysis.
Figures


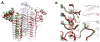





References
-
- Farr RW. Leptospirosis. Clin Infect Dis. 1995;21:1–6. - PubMed
-
- Ren SX, Fu G, Jiang XG, Zeng R, Miao YG, Xu H, Zhang YX, Xiong H, Lu G, Lu LF, Jiang HQ, Jia J, Tu YF, Jiang JX, Gu WY, Zhang YQ, Cai Z, Sheng HH, Yin HF, Zhang Y, Zhu GF, Wan M, Huang HL, Qian Z, Wang SY, Ma W, Yao ZJ, Shen Y, Qiang BQ, Xia QC, Guo XK, Danchin A, Saint Girons I, Somerville RL, Wen YM, Shi MH, Chen Z, Xu JG, Zhao GP. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422:888–893. - PubMed
-
- Bulach DM, Kalambaheti T, de la Pena-Moctezuma A, Adler B. Lipopolysaccharide biosynthesis in Leptospira. J Mol Microbiol Biotechnol. 2000;2:375–380. - PubMed
-
- Weckesser J, Mayer H. Different lipid A types in lipopolysaccharides of phototrophic and related non-phototrophic bacteria. FEMS Microbiol Rev. 1988;4:143–153. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

