A comparative approach to elucidate chloroplast genome replication
- PMID: 19457260
- PMCID: PMC2695485
- DOI: 10.1186/1471-2164-10-237
A comparative approach to elucidate chloroplast genome replication
Abstract
Background: Electron microscopy analyses of replicating chloroplast molecules earlier predicted bidirectional Cairns replication as the prevalent mechanism, perhaps followed by rounds of a rolling circle mechanism. This standard model is being challenged by the recent proposition of homologous recombination-mediated replication in chloroplasts.
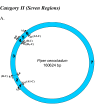
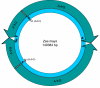
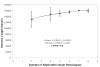
Results: We address this issue in our current study by analyzing nucleotide composition in genome regions between known replication origins, with an aim to reveal any adenine to guanine deamination gradients. These gradual linear gradients typically result from the accumulation of deaminations over the time spent single-stranded by one of the strands of the circular molecule during replication and can, therefore, be used to model the course of replication. Our linear regression analyses on the nucleotide compositions of the non-coding regions and the synonymous third codon position of coding regions, between pairs of replication origins, reveal the existence of significant adenine to guanine deamination gradients in portions overlapping the Small Single Copy (SSC) and the Large Single Copy (LSC) regions between inverted repeats. These gradients increase bi-directionally from the center of each region towards the respective ends, suggesting that both the strands were left single-stranded during replication.
Conclusion: Single-stranded regions of the genome and gradients in time that these regions are left single-stranded, as revealed by our nucleotide composition analyses, appear to converge with the original bi-directional dual displacement loop model and restore evidence for its existence as the primary mechanism. Other proposed faster modes such as homologous recombination and rolling circle initiation could exist in addition to this primary mechanism to facilitate homoplasmy among the intra-cellular chloroplast population.
Figures











Similar articles
-
Analysis of the tobacco chloroplast DNA replication origin (oriB) downstream of the 23 S rRNA gene.J Mol Biol. 1997 May 2;268(2):273-83. doi: 10.1006/jmbi.1997.0972. J Mol Biol. 1997. PMID: 9159470
-
Comparative genomics of four Liliales families inferred from the complete chloroplast genome sequence of Veratrum patulum O. Loes. (Melanthiaceae).Gene. 2013 Nov 10;530(2):229-35. doi: 10.1016/j.gene.2013.07.100. Epub 2013 Aug 23. Gene. 2013. PMID: 23973725
-
Fine mapping of replication origins (ori A and ori B) in Nicotiana tabacum chloroplast DNA.Nucleic Acids Res. 1997 Sep 15;25(18):3681-6. doi: 10.1093/nar/25.18.3681. Nucleic Acids Res. 1997. PMID: 9278490 Free PMC article.
-
Characterization of replication origins flanking the 23S rRNA gene in tobacco chloroplast DNA.Plant Mol Biol. 1996 Nov;32(4):693-706. doi: 10.1007/BF00020210. Plant Mol Biol. 1996. PMID: 8980521
-
Phylogenetic studies and comparative chloroplast genome analyses elucidate the basal position of halophyte Nitraria sibirica (Nitrariaceae) in the Sapindales.Mitochondrial DNA A DNA Mapp Seq Anal. 2018 Jul;29(5):745-755. doi: 10.1080/24701394.2017.1350954. Epub 2017 Jul 15. Mitochondrial DNA A DNA Mapp Seq Anal. 2018. PMID: 28712318
Cited by
-
Theoretical minimal RNA rings designed according to coding constraints mimic deamination gradients.Naturwissenschaften. 2019 Jul 2;106(7-8):44. doi: 10.1007/s00114-019-1638-5. Naturwissenschaften. 2019. PMID: 31267209
-
Variable presence of the inverted repeat and plastome stability in Erodium.Ann Bot. 2016 Jun;117(7):1209-20. doi: 10.1093/aob/mcw065. Epub 2016 Apr 28. Ann Bot. 2016. PMID: 27192713 Free PMC article.
-
Mini-synplastomes for plastid genetic engineering.Plant Biotechnol J. 2022 Feb;20(2):360-373. doi: 10.1111/pbi.13717. Epub 2021 Oct 24. Plant Biotechnol J. 2022. PMID: 34585834 Free PMC article.
-
DNA maintenance in plastids and mitochondria of plants.Front Plant Sci. 2015 Oct 29;6:883. doi: 10.3389/fpls.2015.00883. eCollection 2015. Front Plant Sci. 2015. PMID: 26579143 Free PMC article. Review.
-
The chloroplast genome of the green alga Schizomeris leibleinii (Chlorophyceae) provides evidence for bidirectional DNA replication from a single origin in the chaetophorales.Genome Biol Evol. 2011;3:505-15. doi: 10.1093/gbe/evr037. Epub 2011 May 5. Genome Biol Evol. 2011. PMID: 21546564 Free PMC article.
References
-
- Neuhaus HE, Emes MJ. Nonphotosynthetic Metabolism in Plastids. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:111–140. - PubMed
-
- Maliga P. Engineering the plastid genome of higher plants. Curr Opin Plant Biol. 2002;5:164–172. - PubMed
-
- Kolodner R, Tewari KK. Presence of displacement loops in the covalently closed circular chloroplast deoxyribonucleic acid from higher plants. J Biol Chem. 1975;250:8840–8847. - PubMed
-
- Cairns J. The Form and Duplication of DNA. Endeavour. 1963;22:141–145. - PubMed
-
- Richards OC, Manning JE, Eds . Les Cycles Cellulaires. Editions du CNRS; 1975.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

