Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family
- PMID: 19457860
- PMCID: PMC2742852
- DOI: 10.1074/jbc.M109.002923
Interferon-lambda is functionally an interferon but structurally related to the interleukin-10 family
Abstract
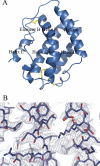
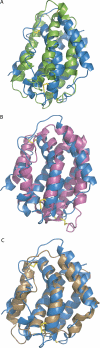
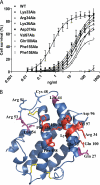
Interferon-lambda (IFN-lambda) is an antiviral cytokine that signals through a distinct receptor complex, composed of the IFN-lambdaR1 and interleukin-10R2 (IL-10R2) receptor chains. We have determined the crystal structure of human IFN-lambda3 and characterized the interaction with its receptor complex through structure-based site-directed mutagenesis. The ability of IFN-lambda3 mutants to signal was determined by measuring the antiviral activity and induced STAT2 phosphorylation. In conclusion, our data show that, although IFN-lambda is functionally an interferon, it is clearly structurally related to members of the IL-10 family. In particular, we found an interesting similarity between IFN-lambda and IL-22, and we suggest that IFN-lambda and IL-22 possess parallel functions, protecting epithelial tissue against viral and bacterial infections, respectively.
Figures


References
-
- Kotenko S. V., Gallagher G., Baurin V. V., Lewis-Antes A., Shen M., Shah N. K., Langer J. A., Sheikh F., Dickensheets H., Donnelly R. P. (2003) Nat. Immunol. 4, 69–77 - PubMed
-
- Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T. E., Kuestner R., Garrigues U., Birks C., Roraback J., Ostrander C., Dong D., Shin J., Presnell S., Fox B., Haldeman B., Cooper E., Taft D., Gilbert T., Grant F. J., Tackett M., Krivan W., McKnight G., Clegg C., Foster D., Klucher K. M. (2003) Nat. Immunol. 4, 63–68 - PubMed
-
- Meager A., Visvalingam K., Dilger P., Bryan D., Wadhwa M. (2005) Cytokine 31, 109–118 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

